Two classes of broadly neutralizing antibodies within a single lineage directed to the high-mannose patch of HIV envelope
- PMID: 25378488
- PMCID: PMC4300629
- DOI: 10.1128/JVI.02905-14
Two classes of broadly neutralizing antibodies within a single lineage directed to the high-mannose patch of HIV envelope
Erratum in
-
Correction for Doores et al., Two Classes of Broadly Neutralizing Antibodies within a Single Lineage Directed to the High-Mannose Patch of HIV Envelope.J Virol. 2015 Jun;89(12):6525. doi: 10.1128/JVI.00593-15. J Virol. 2015. PMID: 25979983 Free PMC article. No abstract available.
Abstract
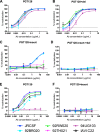
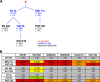
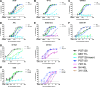
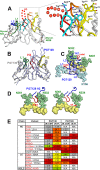
The high-mannose patch of human immunodeficiency virus (HIV) envelope (Env) elicits broadly neutralizing antibodies (bnAbs) during natural infection relatively frequently, and consequently, this region has become a major target of vaccine design. However, it has also become clear that antibody recognition of the region is complex due, at least in part, to variability in neighboring loops and glycans critical to the epitopes. bnAbs against this region have some shared features and some distinguishing features that are crucial to understand in order to design optimal immunogens that can induce different classes of bnAbs against this region. Here, we compare two branches of a single antibody lineage, in which all members recognize the high-mannose patch. One branch (prototype bnAb PGT128) has a 6-amino-acid insertion in CDRH2 that is crucial for broad neutralization. Antibodies in this branch appear to favor a glycan site at N332 on gp120, and somatic hypermutation is required to accommodate the neighboring V1 loop glycans and glycan heterogeneity. The other branch (prototype bnAb PGT130) lacks the CDRH2 insertion. Antibodies in this branch are noticeably effective at neutralizing viruses with an alternate N334 glycan site but are less able to accommodate glycan heterogeneity. We identify a new somatic variant within this branch that is predominantly dependent on N334. The crystal structure of PGT130 offers insight into differences from PGT128. We conclude that different immunogens may be required to elicit bnAbs that have the optimal characteristics of the two branches of the lineage described.
Importance: Development of an HIV vaccine is of vital importance for prevention of new infections, and it is thought that elicitation of HIV bnAbs will be an important component of an effective vaccine. Increasingly, bnAbs that bind to the cluster of high-mannose glycans on the HIV envelope glycoprotein, gp120, are being highlighted as important templates for vaccine design. In particular, bnAbs from IAVI donor 36 (PGT125 to PGT131) have been shown to be extremely broad and potent. Combination of these bnAbs enhanced neutralization breadth considerably, suggesting that an optimal immunogen should elicit several antibodies from this family. Here we study the evolution of this antibody family to inform immunogen design. We identify two classes of bnAbs that differ in their recognition of the high-mannose patch and show that different immunogens may be required to elicit these different classes.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- Kong L, Lee JH, Doores KJ, Murin CD, Julien JP, McBride R, Liu Y, Marozsan A, Cupo A, Klasse PJ, Hoffenberg S, Caulfield M, King CR, Hua Y, Le KM, Khayat R, Deller MC, Clayton T, Tien H, Feizi T, Sanders RW, Paulson JC, Moore JP, Stanfield RL, Burton DR, Ward AB, Wilson IA. 2013. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol 20:796–803. doi: 10.1038/nsmb.2594. - DOI - PMC - PubMed
-
- Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PN, Spencer DI, Seaman MS, Schuitemaker H, Feizi T, Nussenzweig MC, Bjorkman PJ. 2012. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A 109:E3268–E3277. doi: 10.1073/pnas.1217207109. - DOI - PMC - PubMed
-
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. - DOI - PMC - PubMed
-
- Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. 2010. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog 6:e1001028. doi: 10.1371/journal.ppat.1001028. - DOI - PMC - PubMed
-
- Moore PL, Gray ES, Wibmer CK, Bhiman JN, Nonyane M, Sheward DJ, Hermanus T, Bajimaya S, Tumba NL, Abrahams MR, Lambson BE, Ranchobe N, Ping L, Ngandu N, Abdool Karim Q, Abdool Karim SS, Swanstrom RI, Seaman MS, Williamson C, Morris L. 2012. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med 18:1688–1692. doi: 10.1038/nm.2985. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

