Activation of the chicken type I interferon response by infectious bronchitis coronavirus
- PMID: 25378498
- PMCID: PMC4300645
- DOI: 10.1128/JVI.02671-14
Activation of the chicken type I interferon response by infectious bronchitis coronavirus
Abstract
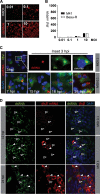
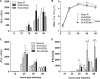
Coronaviruses from both the Alphacoronavirus and Betacoronavirus genera interfere with the type I interferon (IFN) response in various ways, ensuring the limited activation of the IFN response in most cell types. Of the gammacoronaviruses that mainly infect birds, little is known about the activation of the host immune response. We show that the prototypical Gammacoronavirus, infectious bronchitis virus (IBV), induces a delayed activation of the IFN response in primary renal cells, tracheal epithelial cells, and a chicken cell line. In fact, Ifnβ expression is delayed with respect to the peak of viral replication and the accompanying accumulation of double-stranded RNA (dsRNA). In addition, we demonstrate that MDA5 is the primary sensor for Gammacoronavirus infections in chicken cells. Furthermore, we provide evidence that accessory proteins 3a and 3b of IBV modulate the response at the transcriptional and translational levels. Finally, we show that, despite the lack of activation of the IFN response during the early phase of IBV infection, the signaling of nonself dsRNA through both MDA5 and TLR3 remains intact in IBV-infected cells. Taken together, this study provides the first comprehensive analysis of host-virus interactions of a Gammacoronavirus with avian innate immune responses.
Importance: Our results demonstrate that IBV has evolved multiple strategies to avoid the activation of the type I interferon response. Taken together, the present study closes a gap in the understanding of host-IBV interaction and paves the way for further characterization of the mechanisms underlying immune evasion strategies as well as the pathogenesis of gammacoronaviruses.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures





References
-
- Thiel V. 2007. Coronaviruses: molecular and cellular biology. Caister Academic Press, Norfolk, UK.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

