Chromatin landscape and circadian dynamics: Spatial and temporal organization of clock transcription
- PMID: 25378702
- PMCID: PMC4460512
- DOI: 10.1073/pnas.1411264111
Chromatin landscape and circadian dynamics: Spatial and temporal organization of clock transcription
Abstract
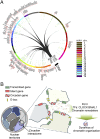
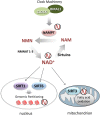
Circadian rhythms drive the temporal organization of a wide variety of physiological and behavioral functions in ∼24-h cycles. This control is achieved through a complex program of gene expression. In mammals, the molecular clock machinery consists of interconnected transcriptional-translational feedback loops that ultimately ensure the proper oscillation of thousands of genes in a tissue-specific manner. To achieve circadian transcriptional control, chromatin remodelers serve the clock machinery by providing appropriate oscillations to the epigenome. Recent findings have revealed the presence of circadian interactomes, nuclear "hubs" of genome topology where coordinately expressed circadian genes physically interact in a spatial and temporal-specific manner. Thus, a circadian nuclear landscape seems to exist, whose interplay with metabolic pathways and clock regulators translates into specific transcriptional programs. Deciphering the molecular mechanisms that connect the circadian clock machinery with the nuclear landscape will reveal yet unexplored pathways that link cellular metabolism to epigenetic control.
Keywords: chromatin; circadian rhythms; epigenetics; nuclear organization.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

