Phenotypic and proteomic characteristics of human dental pulp derived mesenchymal stem cells from a natal, an exfoliated deciduous, and an impacted third molar tooth
- PMID: 25379041
- PMCID: PMC4212660
- DOI: 10.1155/2014/457059
Phenotypic and proteomic characteristics of human dental pulp derived mesenchymal stem cells from a natal, an exfoliated deciduous, and an impacted third molar tooth
Abstract
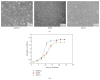
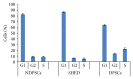
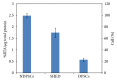
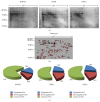
The level of heterogeneity among the isolated stem cells makes them less valuable for clinical use. The purpose of this study was to understand the level of heterogeneity among human dental pulp derived mesenchymal stem cells by using basic cell biology and proteomic approaches. The cells were isolated from a natal (NDPSCs), an exfoliated deciduous (stem cells from human exfoliated deciduous (SHED)), and an impacted third molar (DPSCs) tooth of three different donors. All three stem cells displayed similar features related to morphology, proliferation rates, expression of various cell surface markers, and differentiation potentials into adipocytes, osteocytes, and chondrocytes. Furthermore, using 2DE approach coupled with MALDI-TOF/TOF, we have generated a common 2DE profile for all three stem cells. We found that 62.3 ± 7% of the protein spots were conserved among the three mesenchymal stem cell lines. Sixty-one of these conserved spots were identified by MALDI-TOF/TOF analysis. Classification of the identified proteins based on biological function revealed that structurally important proteins and proteins that are involved in protein folding machinery are predominantly expressed by all three stem cell lines. Some of these proteins may hold importance in understanding specific properties of human dental pulp derived mesenchymal stem cells.
Figures



References
-
- Cookson C. Stem cells: East...and West. Country report: United Kingdom. Scientific American. 2005;293(1, article A23) - PubMed
-
- Wagner W., Feldmann R. E., Jr., Seckinger A., Maurer M. H., Wein F., Blake J., Krause U., Kalenka A., Bürgers H. F., Saffrich R., Wuchter P., Kuschinsky W., Ho A. D. The heterogeneity of human mesenchymal stem cell preparations—evidence from simultaneous analysis of proteomes and transcriptomes. Experimental Hematology. 2006;34(4):536–548. doi: 10.1016/j.exphem.2006.01.002. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

