Random Forest ensembles for detection and prediction of Alzheimer's disease with a good between-cohort robustness
- PMID: 25379423
- PMCID: PMC4215532
- DOI: 10.1016/j.nicl.2014.08.023
Random Forest ensembles for detection and prediction of Alzheimer's disease with a good between-cohort robustness
Abstract
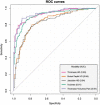
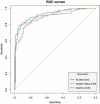
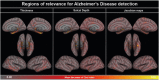
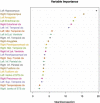
Computer-aided diagnosis of Alzheimer's disease (AD) is a rapidly developing field of neuroimaging with strong potential to be used in practice. In this context, assessment of models' robustness to noise and imaging protocol differences together with post-processing and tuning strategies are key tasks to be addressed in order to move towards successful clinical applications. In this study, we investigated the efficacy of Random Forest classifiers trained using different structural MRI measures, with and without neuroanatomical constraints in the detection and prediction of AD in terms of accuracy and between-cohort robustness. From The ADNI database, 185 AD, and 225 healthy controls (HC) were randomly split into training and testing datasets. 165 subjects with mild cognitive impairment (MCI) were distributed according to the month of conversion to dementia (4-year follow-up). Structural 1.5-T MRI-scans were processed using Freesurfer segmentation and cortical reconstruction. Using the resulting output, AD/HC classifiers were trained. Training included model tuning and performance assessment using out-of-bag estimation. Subsequently the classifiers were validated on the AD/HC test set and for the ability to predict MCI-to-AD conversion. Models' between-cohort robustness was additionally assessed using the AddNeuroMed dataset acquired with harmonized clinical and imaging protocols. In the ADNI set, the best AD/HC sensitivity/specificity (88.6%/92.0% - test set) was achieved by combining cortical thickness and volumetric measures. The Random Forest model resulted in significantly higher accuracy compared to the reference classifier (linear Support Vector Machine). The models trained using parcelled and high-dimensional (HD) input demonstrated equivalent performance, but the former was more effective in terms of computation/memory and time costs. The sensitivity/specificity for detecting MCI-to-AD conversion (but not AD/HC classification performance) was further improved from 79.5%/75%-83.3%/81.3% by a combination of morphometric measurements with ApoE-genotype and demographics (age, sex, education). When applied to the independent AddNeuroMed cohort, the best ADNI models produced equivalent performance without substantial accuracy drop, suggesting good robustness sufficient for future clinical implementation.
Keywords: ADNI; AddNeuroMed; Alzheimer's disease; Computer-aided diagnosis; Mild cognitive impairment; Multi-center study; Random Forest; Structural MRI.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

