Microseconds simulations reveal a new sodium-binding site and the mechanism of sodium-coupled substrate uptake by LeuT
- PMID: 25381247
- PMCID: PMC4281755
- DOI: 10.1074/jbc.M114.617555
Microseconds simulations reveal a new sodium-binding site and the mechanism of sodium-coupled substrate uptake by LeuT
Abstract
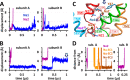
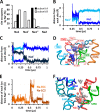
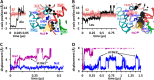
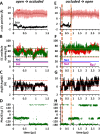
The bacterial sodium-coupled leucine/alanine transporter LeuT is broadly used as a model system for studying the transport mechanism of neurotransmitters because of its structural and functional homology to mammalian transporters such as serotonin, dopamine, or norepinephrine transporters, and because of the resolution of its structure in different states. Although the binding sites (S1 for substrate, and Na1 and Na2 for two co-transported sodium ions) have been resolved, we still lack a mechanistic understanding of coupled Na(+)- and substrate-binding events. We present here results from extensive (>20 μs) unbiased molecular dynamics simulations generated using the latest computing technology. Simulations show that sodium binds initially the Na1 site, but not Na2, and, consistently, sodium unbinding/escape to the extracellular (EC) region first takes place at Na2, succeeded by Na1. Na2 diffusion back to the EC medium requires prior dissociation of substrate from S1. Significantly, Na(+) binding (and unbinding) consistently involves a transient binding to a newly discovered site, Na1″, near S1, as an intermediate state. A robust sequence of substrate uptake events coupled to sodium bindings and translocations between those sites assisted by hydration emerges from the simulations: (i) bindings of a first Na(+) to Na1″, translocation to Na1, a second Na(+) to vacated Na1″ and then to Na2, and substrate to S1; (ii) rotation of Phe(253) aromatic group to seclude the substrate from the EC region; and (iii) concerted tilting of TM1b and TM6a toward TM3 and TM8 to close the EC vestibule.
Keywords: Biophysics; LeuT; Mechanism of Binding; Membrane Transport; NSS; Neurotransmitter Transport; Sodium Transport; Sodium-coupled; Transport.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Richerson G. B., Wu Y. (2004) Role of the GABA transporter in epilepsy. Adv. Exp. Med. Biol. 548, 76–91 - PubMed
-
- Andersen J., Kristensen A. S., Bang-Andersen B., Stromgaard K. (2009) Recent advances in the understanding of the interaction of antidepressant drugs with serotonin and norepinephrine transporters. Chem. Commun. (Camb.) 25, 3677–3692 - PubMed
-
- Clausen R. P., Madsen K., Larsson O. M., Frølund B., Krogsgaard-Larsen P., Schousboe A. (2006) Structure-activity relationship and pharmacology of γ-aminobutyric acid (GABA) transport inhibitors. Adv. Pharmacol. 54, 265–284 - PubMed
-
- Hahn M. K., Blakely R. D. (2002) Monoamine transporter gene structure and polymorphisms in relation to psychiatric and other complex disorders. Pharmacogenomics J. 2, 217–235 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

