Functional classification of BRCA2 DNA variants by splicing assays in a large minigene with 9 exons
- PMID: 25382762
- PMCID: PMC4371643
- DOI: 10.1002/humu.22725
Functional classification of BRCA2 DNA variants by splicing assays in a large minigene with 9 exons
Abstract
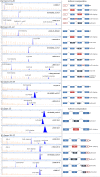
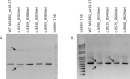
Numerous pathogenic DNA variants impair the splicing mechanism in human genetic diseases. Minigenes are optimal approaches to test variants under the splicing viewpoint without the need of patient samples. We aimed to design a robust minigene construct of the breast cancer gene BRCA2 in order to investigate the impact of variants on splicing. BRCA2 exons 19-27 (MGBR2_ex19-27) were cloned in the new vector pSAD. It produced a large transcript of the expected size (2,174 nucleotides) and exon structure (V1-ex19-27-V2). Splicing assays showed that 18 (17 splice-site and 1 silencer variants) out of 40 candidate DNA variants induced aberrant patterns. Twenty-four anomalous transcripts were accurately detected by fluorescent-RT-PCR that were generated by exon-skipping, alternative site usage, and intron-retention events. Fourteen variants induced major anomalies and were predicted to disrupt protein function so they could be classified as pathogenic. Furthermore, minigene mimicked previously reported patient RNA outcomes of seven variants supporting the reproducibility of minigene assays. Therefore, a relevant fraction of variants are involved in breast cancer through splicing alterations. MGBR2_ex19-27 is the largest reported BRCA2 minigene and constitutes a valuable tool for the functional and clinical classification of sequence variations.
Keywords: BRCA2; hereditary breast and ovarian cancer; minigenes; splicing; unclassified variants.
© 2014 WILEY PERIODICALS, INC.
Figures


References
-
- Baralle M, Skoko N, Knezevich A, De CL, Motti D, Bhuvanagiri M, Baralle D, Buratti E, Baralle FE. NF1 mRNA biogenesis: effect of the genomic milieu in splicing regulation of the NF1 exon 37 region. FEBS Lett. 2006;580:4449–4456. - PubMed
-
- Bonatti F, Pepe C, Tancredi M, Lombardi G, Aretini P, Sensi E, Falaschi E, Cipollini G, Bevilacqua G, Caligo MA. RNA-based analysis of BRCA1 and BRCA2 gene alterations. Cancer Genet Cytogenet. 2006;170:93–101. - PubMed
-
- Bonnet C, Krieger S, Vezain M, Rousselin A, Tournier I, Martins A, Berthet P, Chevrier A, Dugast C, Layet V, Rossi A, Lidereau R, et al. Screening BRCA1 and BRCA2 unclassified variants for splicing mutations using reverse transcription PCR on patient RNA and an ex vivo assay based on a splicing reporter minigene. J Med Genet. 2008;45:438–446. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

