mTORC1-mediated translational elongation limits intestinal tumour initiation and growth
- PMID: 25383520
- PMCID: PMC4304784
- DOI: 10.1038/nature13896
mTORC1-mediated translational elongation limits intestinal tumour initiation and growth
Abstract
Inactivation of APC is a strongly predisposing event in the development of colorectal cancer, prompting the search for vulnerabilities specific to cells that have lost APC function. Signalling through the mTOR pathway is known to be required for epithelial cell proliferation and tumour growth, and the current paradigm suggests that a critical function of mTOR activity is to upregulate translational initiation through phosphorylation of 4EBP1 (refs 6, 7). This model predicts that the mTOR inhibitor rapamycin, which does not efficiently inhibit 4EBP1 (ref. 8), would be ineffective in limiting cancer progression in APC-deficient lesions. Here we show in mice that mTOR complex 1 (mTORC1) activity is absolutely required for the proliferation of Apc-deficient (but not wild-type) enterocytes, revealing an unexpected opportunity for therapeutic intervention. Although APC-deficient cells show the expected increases in protein synthesis, our study reveals that it is translation elongation, and not initiation, which is the rate-limiting component. Mechanistically, mTORC1-mediated inhibition of eEF2 kinase is required for the proliferation of APC-deficient cells. Importantly, treatment of established APC-deficient adenomas with rapamycin (which can target eEF2 through the mTORC1-S6K-eEF2K axis) causes tumour cells to undergo growth arrest and differentiation. Taken together, our data suggest that inhibition of translation elongation using existing, clinically approved drugs, such as the rapalogs, would provide clear therapeutic benefit for patients at high risk of developing colorectal cancer.
Figures
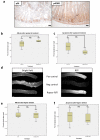
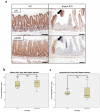
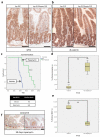
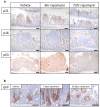






Comment in
-
Cancer: mTOR inhibition curbs colorectal cancer.Nat Rev Drug Discov. 2015 Jan;14(1):14-5. doi: 10.1038/nrd4523. Nat Rev Drug Discov. 2015. PMID: 25549586 No abstract available.
References
-
- Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. - PubMed
-
- Korinek V, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. - PubMed
-
- Fujishita T, Aoki K, Lane HA, Aoki M, Taketo MM. Inhibition of the mTORC1 pathway suppresses intestinal polyp formation and reduces mortality in ApcDelta716 mice. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13544–13549. doi:10.1073/pnas.0800041105. - PMC - PubMed
Methods Reference
-
- El Marjou F, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi:10.1002/gene.20042. - PubMed
-
- Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi:nature06196 [pii] 10.1038/nature06196. - PubMed
-
- Shibata H, et al. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science. 1997;278:120–123. - PubMed
-
- Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. - PubMed
-
- de Alboran IM, et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G1000078/1/NC3RS_/National Centre for the Replacement, Refinement and Reduction of Animals in Research/United Kingdom
- 10-0643/AICR_/Worldwide Cancer Research/United Kingdom
- MC_EX_G0902052/MRC_/Medical Research Council/United Kingdom
- MC_UP_A600_1023/MRC_/Medical Research Council/United Kingdom
- 21139/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

