Discovery of a bacterial 5-methylcytosine deaminase
- PMID: 25384249
- PMCID: PMC4255641
- DOI: 10.1021/bi5012767
Discovery of a bacterial 5-methylcytosine deaminase
Abstract
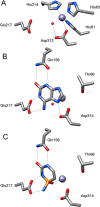
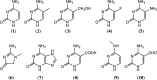
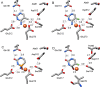
5-Methylcytosine is found in all domains of life, but the bacterial cytosine deaminase from Escherichia coli (CodA) will not accept 5-methylcytosine as a substrate. Since significant amounts of 5-methylcytosine are produced in both prokaryotes and eukaryotes, this compound must eventually be catabolized and the fragments recycled by enzymes that have yet to be identified. We therefore initiated a comprehensive phylogenetic screen for enzymes that may be capable of deaminating 5-methylcytosine to thymine. From a systematic analysis of sequence homologues of CodA from thousands of bacterial species, we identified putative cytosine deaminases where a "discriminating" residue in the active site, corresponding to Asp-314 in CodA from E. coli, was no longer conserved. Representative examples from Klebsiella pneumoniae (locus tag: Kpn00632), Rhodobacter sphaeroides (locus tag: Rsp0341), and Corynebacterium glutamicum (locus tag: NCgl0075) were demonstrated to efficiently deaminate 5-methylcytosine to thymine with values of kcat/Km of 1.4 × 10(5), 2.9 × 10(4), and 1.1 × 10(3) M(-1) s(-1), respectively. These three enzymes also catalyze the deamination of 5-fluorocytosine to 5-fluorouracil with values of kcat/Km of 1.2 × 10(5), 6.8 × 10(4), and 2.0 × 10(2) M(-1) s(-1), respectively. The three-dimensional structure of Kpn00632 was determined by X-ray diffraction methods with 5-methylcytosine (PDB id: 4R85 ), 5-fluorocytosine (PDB id: 4R88 ), and phosphonocytosine (PDB id: 4R7W ) bound in the active site. When thymine auxotrophs of E. coli express these enzymes, they are capable of growth in media lacking thymine when supplemented with 5-methylcytosine. Expression of these enzymes in E. coli is toxic in the presence of 5-fluorocytosine, due to the efficient transformation to 5-fluorouracil.
Figures
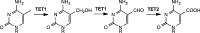




Similar articles
-
Alanine-scanning mutagenesis reveals a cytosine deaminase mutant with altered substrate preference.Biochemistry. 2004 Jul 20;43(28):8957-64. doi: 10.1021/bi049720z. Biochemistry. 2004. PMID: 15248753
-
N4-Methylcytosine Supports the Growth of Escherichia coli Uracil Auxotrophs.Int J Mol Sci. 2025 Feb 20;26(5):1812. doi: 10.3390/ijms26051812. Int J Mol Sci. 2025. PMID: 40076440 Free PMC article.
-
The structure of Escherichia coli cytosine deaminase.J Mol Biol. 2002 Jan 25;315(4):687-97. doi: 10.1006/jmbi.2001.5277. J Mol Biol. 2002. PMID: 11812140
-
Synthesis and characterization of a novel chitosan based E. coli cytosine deaminase nanocomposite for potential application in prodrug enzyme therapy.Biotechnol Lett. 2011 Jan;33(1):153-7. doi: 10.1007/s10529-010-0416-4. Epub 2010 Oct 20. Biotechnol Lett. 2011. PMID: 20960222
-
Emerging implications of nonmammalian cytosine deaminases on cancer therapeutics.Appl Biochem Biotechnol. 2012 Aug;167(7):2103-16. doi: 10.1007/s12010-012-9746-0. Epub 2012 Jun 7. Appl Biochem Biotechnol. 2012. PMID: 22673971 Review.
Cited by
-
Structural characterization of an isocytosine-specific deaminase VCZ reveals its application potential in the anti-cancer therapy.iScience. 2023 Aug 18;26(9):107672. doi: 10.1016/j.isci.2023.107672. eCollection 2023 Sep 15. iScience. 2023. PMID: 37680460 Free PMC article.
-
RudS: bacterial desulfidase responsible for tRNA 4-thiouridine de-modification.Nucleic Acids Res. 2024 Sep 23;52(17):10543-10562. doi: 10.1093/nar/gkae716. Nucleic Acids Res. 2024. PMID: 39166491 Free PMC article.
-
Structural Determinants for Substrate Selectivity in Guanine Deaminase Enzymes of the Amidohydrolase Superfamily.Biochemistry. 2019 Jul 30;58(30):3280-3292. doi: 10.1021/acs.biochem.9b00341. Epub 2019 Jul 19. Biochemistry. 2019. PMID: 31283204 Free PMC article.
-
Nucleobase deaminases: a potential enzyme system for new therapies.RSC Adv. 2018 Jun 28;8(42):23567-23577. doi: 10.1039/c8ra04112a. eCollection 2018 Jun 27. RSC Adv. 2018. PMID: 35540270 Free PMC article. Review.
-
Discovery of Bacterial Deaminases That Convert 5-Fluoroisocytosine Into 5-Fluorouracil.Front Microbiol. 2018 Oct 8;9:2375. doi: 10.3389/fmicb.2018.02375. eCollection 2018. Front Microbiol. 2018. PMID: 30349513 Free PMC article.
References
-
- Suzuki M. M.; Bird A. (2008) DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 9, 465–476. - PubMed
-
- Bestor T.; Laudano A.; Mattaliano R.; Ingram V. (1988) Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells: The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol. 203, 971–983. - PubMed
-
- Pavlopoulou A.; Kossida S. (2007) Plant cytosine-5 DNA methyltransferases: Structure, function, and molecular evolution. Genomics 90, 530–541. - PubMed
-
- Jabbari K.; Bernardi G. (2004) Cytosine methylation and CpG, TpG (CpA) and TpA frequencies. Gene 333, 143–149. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

