Discovery of a bacterial 5-methylcytosine deaminase
- PMID: 25384249
- PMCID: PMC4255641
- DOI: 10.1021/bi5012767
Discovery of a bacterial 5-methylcytosine deaminase
Abstract
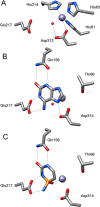
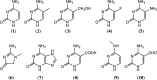
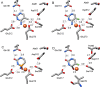
5-Methylcytosine is found in all domains of life, but the bacterial cytosine deaminase from Escherichia coli (CodA) will not accept 5-methylcytosine as a substrate. Since significant amounts of 5-methylcytosine are produced in both prokaryotes and eukaryotes, this compound must eventually be catabolized and the fragments recycled by enzymes that have yet to be identified. We therefore initiated a comprehensive phylogenetic screen for enzymes that may be capable of deaminating 5-methylcytosine to thymine. From a systematic analysis of sequence homologues of CodA from thousands of bacterial species, we identified putative cytosine deaminases where a "discriminating" residue in the active site, corresponding to Asp-314 in CodA from E. coli, was no longer conserved. Representative examples from Klebsiella pneumoniae (locus tag: Kpn00632), Rhodobacter sphaeroides (locus tag: Rsp0341), and Corynebacterium glutamicum (locus tag: NCgl0075) were demonstrated to efficiently deaminate 5-methylcytosine to thymine with values of kcat/Km of 1.4 × 10(5), 2.9 × 10(4), and 1.1 × 10(3) M(-1) s(-1), respectively. These three enzymes also catalyze the deamination of 5-fluorocytosine to 5-fluorouracil with values of kcat/Km of 1.2 × 10(5), 6.8 × 10(4), and 2.0 × 10(2) M(-1) s(-1), respectively. The three-dimensional structure of Kpn00632 was determined by X-ray diffraction methods with 5-methylcytosine (PDB id: 4R85 ), 5-fluorocytosine (PDB id: 4R88 ), and phosphonocytosine (PDB id: 4R7W ) bound in the active site. When thymine auxotrophs of E. coli express these enzymes, they are capable of growth in media lacking thymine when supplemented with 5-methylcytosine. Expression of these enzymes in E. coli is toxic in the presence of 5-fluorocytosine, due to the efficient transformation to 5-fluorouracil.
Figures
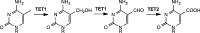




References
-
- Suzuki M. M.; Bird A. (2008) DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 9, 465–476. - PubMed
-
- Bestor T.; Laudano A.; Mattaliano R.; Ingram V. (1988) Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells: The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol. 203, 971–983. - PubMed
-
- Pavlopoulou A.; Kossida S. (2007) Plant cytosine-5 DNA methyltransferases: Structure, function, and molecular evolution. Genomics 90, 530–541. - PubMed
-
- Jabbari K.; Bernardi G. (2004) Cytosine methylation and CpG, TpG (CpA) and TpA frequencies. Gene 333, 143–149. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

