Human CHD2 is a chromatin assembly ATPase regulated by its chromo- and DNA-binding domains
- PMID: 25384982
- PMCID: PMC4281729
- DOI: 10.1074/jbc.M114.609156
Human CHD2 is a chromatin assembly ATPase regulated by its chromo- and DNA-binding domains
Abstract
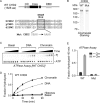
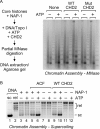
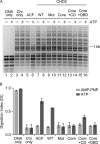
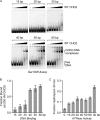
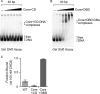
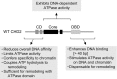
Chromodomain helicase DNA-binding protein 2 (CHD2) is an ATPase and a member of the SNF2-like family of helicase-related enzymes. Although deletions of CHD2 have been linked to developmental defects in mice and epileptic disorders in humans, little is known about its biochemical and cellular activities. In this study, we investigate the ATP-dependent activity of CHD2 and show that CHD2 catalyzes the assembly of chromatin into periodic arrays. We also show that the N-terminal region of CHD2, which contains tandem chromodomains, serves an auto-inhibitory role in both the DNA-binding and ATPase activities of CHD2. While loss of the N-terminal region leads to enhanced chromatin-stimulated ATPase activity, the N-terminal region is required for ATP-dependent chromatin remodeling by CHD2. In contrast, the C-terminal region, which contains a putative DNA-binding domain, selectively senses double-stranded DNA of at least 40 base pairs in length and enhances the ATPase and chromatin remodeling activities of CHD2. Our study shows that the accessory domains of CHD2 play central roles in both regulating the ATPase domain and conferring selectivity to chromatin substrates.
Keywords: ATPase; CHD2; Chromatin Remodeling; Chromodomains; DNA-binding Protein; Epilepsy; Nucleosome.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
-
- Gorbalenya A. E., Koonin E. V. (1993) Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3, 419–429
-
- Clapier C. R., Cairns B. R. (2009) The Biology of Chromatin Remodeling Complexes. Annu. Rev. Biochem. 78, 273–304 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

