Proteotoxic crisis, the ubiquitin-proteasome system, and cancer therapy
- PMID: 25385277
- PMCID: PMC4226866
- DOI: 10.1186/s12915-014-0094-0
Proteotoxic crisis, the ubiquitin-proteasome system, and cancer therapy
Abstract
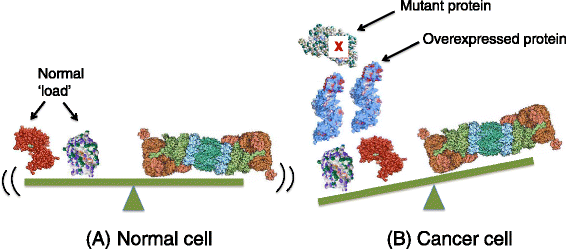
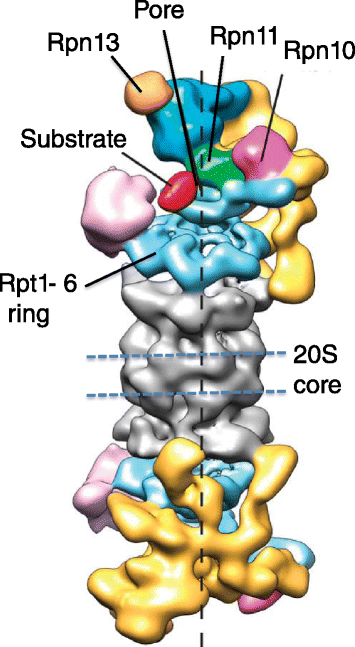
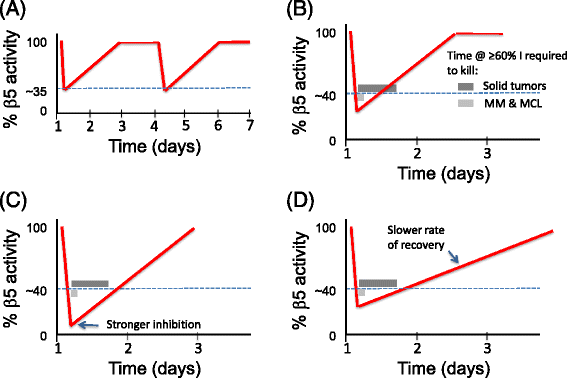
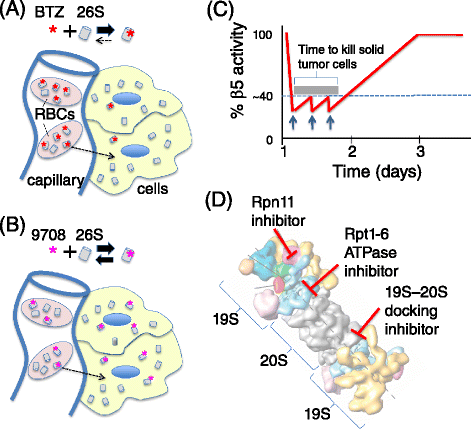
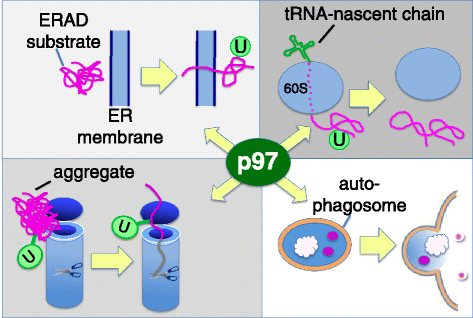
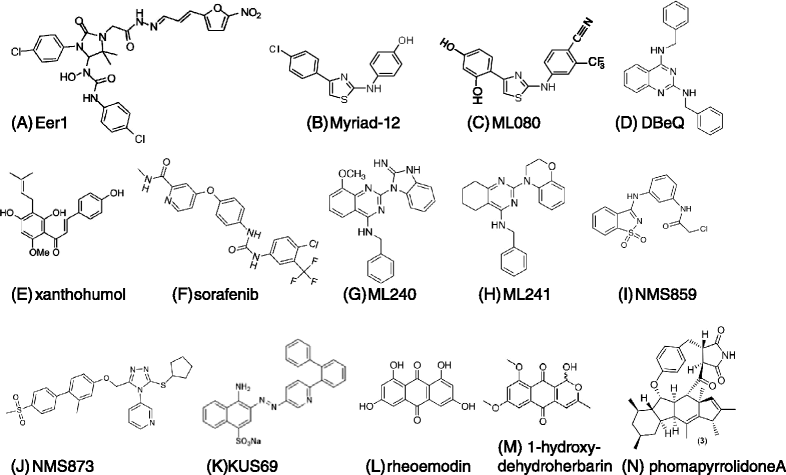
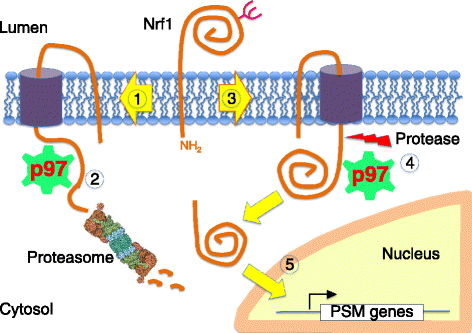
Genomic alterations may make cancer cells more dependent than normal cells on mechanisms of proteostasis, including protein folding and degradation. This proposition is the basis for the clinical use of proteasome inhibitors to treat multiple myeloma and mantle cell lymphoma. However, proteasome inhibitors have not proved effective in treating other cancers, and this has called into question the general applicability of this approach. Here, I consider possible explanations for this apparently limited applicability, and discuss whether inhibiting other broadly acting components of the ubiquitin-proteasome system - including ubiquitin-activating enzyme and the AAA-ATPase p97/VCP - might be more generally effective in cancer therapy.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

