Prediction of interindividual differences in hepatic functions and drug sensitivity by using human iPS-derived hepatocytes
- PMID: 25385620
- PMCID: PMC4250156
- DOI: 10.1073/pnas.1413481111
Prediction of interindividual differences in hepatic functions and drug sensitivity by using human iPS-derived hepatocytes
Abstract
Interindividual differences in hepatic metabolism, which are mainly due to genetic polymorphism in its gene, have a large influence on individual drug efficacy and adverse reaction. Hepatocyte-like cells (HLCs) differentiated from human induced pluripotent stem (iPS) cells have the potential to predict interindividual differences in drug metabolism capacity and drug response. However, it remains uncertain whether human iPSC-derived HLCs can reproduce the interindividual difference in hepatic metabolism and drug response. We found that cytochrome P450 (CYP) metabolism capacity and drug responsiveness of the primary human hepatocytes (PHH)-iPS-HLCs were highly correlated with those of PHHs, suggesting that the PHH-iPS-HLCs retained donor-specific CYP metabolism capacity and drug responsiveness. We also demonstrated that the interindividual differences, which are due to the diversity of individual SNPs in the CYP gene, could also be reproduced in PHH-iPS-HLCs. We succeeded in establishing, to our knowledge, the first PHH-iPS-HLC panel that reflects the interindividual differences of hepatic drug-metabolizing capacity and drug responsiveness.
Keywords: CYP2D6; SNP; hepatocyte; human iPS cells; personalized drug therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
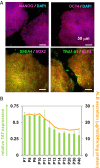
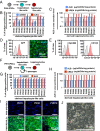
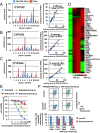

References
-
- Ingelman-Sundberg M. Pharmacogenetics of cytochrome P450 and its applications in drug therapy: The past, present and future. Trends Pharmacol Sci. 2004;25(4):193–200. - PubMed
-
- Ingelman-Sundberg M. Genetic susceptibility to adverse effects of drugs and environmental toxicants. The role of the CYP family of enzymes. Mutat Res. 2001;482(1-2):11–19. - PubMed
-
- Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: Part I. Clin Pharmacokinet. 2009;48(11):689–723. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

