NET23/STING promotes chromatin compaction from the nuclear envelope
- PMID: 25386906
- PMCID: PMC4227661
- DOI: 10.1371/journal.pone.0111851
NET23/STING promotes chromatin compaction from the nuclear envelope
Abstract
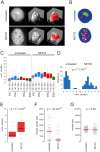
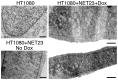
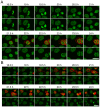
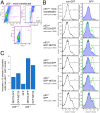
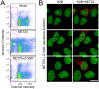
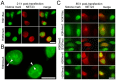
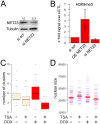
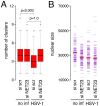
Changes in the peripheral distribution and amount of condensed chromatin are observed in a number of diseases linked to mutations in the lamin A protein of the nuclear envelope. We postulated that lamin A interactions with nuclear envelope transmembrane proteins (NETs) that affect chromatin structure might be altered in these diseases and so screened thirty-one NETs for those that promote chromatin compaction as determined by an increase in the number of chromatin clusters of high pixel intensity. One of these, NET23 (also called STING, MITA, MPYS, ERIS, Tmem173), strongly promoted chromatin compaction. A correlation between chromatin compaction and endogenous levels of NET23/STING was observed for a number of human cell lines, suggesting that NET23/STING may contribute generally to chromatin condensation. NET23/STING has separately been found to be involved in innate immune response signaling. Upon infection cells make a choice to either apoptose or to alter chromatin architecture to support focused expression of interferon genes and other response factors. We postulate that the chromatin compaction induced by NET23/STING may contribute to this choice because the cells expressing NET23/STING eventually apoptose, but the chromatin compaction effect is separate from this as the condensation was still observed when cells were treated with Z-VAD to block apoptosis. NET23/STING-induced compacted chromatin revealed changes in epigenetic marks including changes in histone methylation and acetylation. This indicates a previously uncharacterized nuclear role for NET23/STING potentially in both innate immune signaling and general chromatin architecture.
Conflict of interest statement
Figures




Similar articles
-
Condensins exert force on chromatin-nuclear envelope tethers to mediate nucleoplasmic reticulum formation in Drosophila melanogaster.G3 (Bethesda). 2014 Dec 30;5(3):341-52. doi: 10.1534/g3.114.015685. G3 (Bethesda). 2014. PMID: 25552604 Free PMC article.
-
The leukocyte nuclear envelope proteome varies with cell activation and contains novel transmembrane proteins that affect genome architecture.Mol Cell Proteomics. 2010 Dec;9(12):2571-85. doi: 10.1074/mcp.M110.002915. Epub 2010 Aug 6. Mol Cell Proteomics. 2010. PMID: 20693407 Free PMC article.
-
Histones and histone modifications in perinuclear chromatin anchoring: from yeast to man.EMBO Rep. 2016 Feb;17(2):139-55. doi: 10.15252/embr.201541809. Epub 2016 Jan 20. EMBO Rep. 2016. PMID: 26792937 Free PMC article. Review.
-
DNA methylation affects nuclear organization, histone modifications, and linker histone binding but not chromatin compaction.J Cell Biol. 2007 May 7;177(3):401-11. doi: 10.1083/jcb.200607133. J Cell Biol. 2007. PMID: 17485486 Free PMC article.
-
Nuclear envelope proteins and chromatin arrangement: a pathogenic mechanism for laminopathies.Eur J Histochem. 2006 Jan-Mar;50(1):1-8. Eur J Histochem. 2006. PMID: 16584978 Review.
Cited by
-
STING protects breast cancer cells from intrinsic and genotoxic-induced DNA instability via a non-canonical, cell-autonomous pathway.Oncogene. 2021 Dec;40(49):6627-6640. doi: 10.1038/s41388-021-02037-4. Epub 2021 Oct 8. Oncogene. 2021. PMID: 34625708
-
Intrinsic antiproliferative activity of the innate sensor STING in T lymphocytes.J Exp Med. 2017 Jun 5;214(6):1769-1785. doi: 10.1084/jem.20161674. Epub 2017 May 8. J Exp Med. 2017. PMID: 28484079 Free PMC article.
-
Gene expression profiling of CD34(+) cells from patients with myeloproliferative neoplasms.Oncol Lett. 2021 Mar;21(3):204. doi: 10.3892/ol.2021.12465. Epub 2021 Jan 12. Oncol Lett. 2021. PMID: 33574943 Free PMC article.
-
In silico synchronization reveals regulators of nuclear ruptures in lamin A/C deficient model cells.Sci Rep. 2016 Jul 27;6:30325. doi: 10.1038/srep30325. Sci Rep. 2016. PMID: 27461848 Free PMC article.
-
Innate immunity mediator STING modulates nascent DNA metabolism at stalled forks in human cells.Front Mol Biosci. 2023 Jan 12;9:1048726. doi: 10.3389/fmolb.2022.1048726. eCollection 2022. Front Mol Biosci. 2023. PMID: 36710880 Free PMC article.
References
-
- Maraldi NM, Capanni C, Cenni V, Fini M, Lattanzi G (2011) Laminopathies and lamin-associated signaling pathways. J Cell Biochem 112: 979–992. - PubMed
-
- Sewry CA, Brown SC, Mercuri E, Bonne G, Feng L, et al. (2001) Skeletal muscle pathology in autosomal dominant Emery-Dreifuss muscular dystrophy with lamin A/C mutations. Neuropathol Appl Neurobiol 27: 281–290. - PubMed
-
- Verga L, Concardi M, Pilotto A, Bellini O, Pasotti M, et al. (2003) Loss of lamin A/C expression revealed by immuno-electron microscopy in dilated cardiomyopathy with atrioventricular block caused by LMNA gene defects. Virchows Arch 443: 664–671. - PubMed
-
- Fidzianska A, Toniolo D, Hausmanowa-Petrusewicz I (1998) Ultrastructural abnormality of sarcolemmal nuclei in Emery-Dreifuss muscular dystrophy (EDMD). J Neurol Sci 159: 88–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

