Suppression of the FOXM1 transcriptional programme via novel small molecule inhibition
- PMID: 25387393
- PMCID: PMC4258842
- DOI: 10.1038/ncomms6165
Suppression of the FOXM1 transcriptional programme via novel small molecule inhibition
Abstract
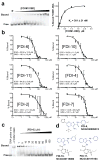
The transcription factor FOXM1 binds to sequence-specific motifs on DNA (C/TAAACA) through its DNA-binding domain (DBD) and activates proliferation- and differentiation-associated genes. Aberrant overexpression of FOXM1 is a key feature in oncogenesis and progression of many human cancers. Here--from a high-throughput screen applied to a library of 54,211 small molecules--we identify novel small molecule inhibitors of FOXM1 that block DNA binding. One of the identified compounds, FDI-6 (NCGC00099374), is characterized in depth and is shown to bind directly to FOXM1 protein, to displace FOXM1 from genomic targets in MCF-7 breast cancer cells, and induce concomitant transcriptional downregulation. Global transcript profiling of MCF-7 cells by RNA-seq shows that FDI-6 specifically downregulates FOXM1-activated genes with FOXM1 occupancy confirmed by ChIP-PCR. This small molecule-mediated effect is selective for FOXM1-controlled genes with no effect on genes regulated by homologous forkhead family factors.
Conflict of interest statement
Figures




References
-
- Darnell JE. Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. - PubMed
-
- Libermann TA, Zerbini LF. Targeting transcription factors for cancer gene therapy. Curr Gene Ther. 2006;6:17–33. - PubMed
-
- Laoukili J, Stahl M, Medema RH. FoxM1: At the crossroads of ageing and cancer. Biochimica et Biophysica Acta (BBA)- Reviews on Cancer. 2007;1775:92–102. - PubMed
-
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nature Reviews Genetics. 2009;10:252–263. - PubMed
-
- Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nature Reviews Genetics. 2012;13:613–626. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

