Expression of MTAP inhibits tumor-related phenotypes in HT1080 cells via a mechanism unrelated to its enzymatic function
- PMID: 25387827
- PMCID: PMC4291467
- DOI: 10.1534/g3.114.014555
Expression of MTAP inhibits tumor-related phenotypes in HT1080 cells via a mechanism unrelated to its enzymatic function
Abstract
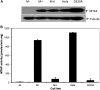
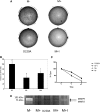
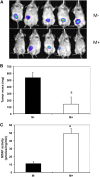
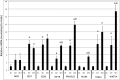
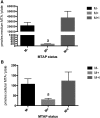
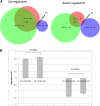
Methylthioadenosine Phosphorylase (MTAP) is a tumor suppressor gene that is frequently deleted in human cancers and encodes an enzyme responsible for the catabolism of the polyamine byproduct 5'deoxy-5'-methylthioadenosine (MTA). To elucidate the mechanism by which MTAP inhibits tumor formation, we have reintroduced MTAP into MTAP-deleted HT1080 fibrosarcoma cells. Expression of MTAP resulted in a variety of phenotypes, including decreased colony formation in soft-agar, decreased migration, decreased in vitro invasion, increased matrix metalloproteinase production, and reduced ability to form tumors in severe combined immunodeficiency mice. Microarray analysis showed that MTAP affected the expression of genes involved in a variety of processes, including cell adhesion, extracellular matrix interaction, and cell signaling. Treatment of MTAP-expressing cells with a potent inhibitor of MTAP's enzymatic activity (MT-DADMe-ImmA) did not result in a MTAP- phenotype. This finding suggests that MTAP's tumor suppressor function is not the same as its known enzymatic function. To confirm this, we introduced a catalytically inactive version of MTAP, D220A, into HT1080 cells and found that this mutant was fully capable of reversing the soft agar colony formation, migration, and matrix metalloproteinase phenotypes. Our results show that MTAP affects cellular phenotypes in HT1080 cells in a manner that is independent of its known enzymatic activity.
Keywords: cell mobility and migration; methionine; suppressor genes.
Copyright © 2015 Tang et al.
Figures
References
-
- Basu I., Cordovano G., Das I., Belbin T. J., Guha C., et al. , 2007. A transition state analogue of 5′-methylthioadenosine phosphorylase induces apoptosis in head and neck cancers. J. Biol. Chem. 282: 21477–21486. - PubMed
-
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc., B 57: 289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
