QseC inhibitors as an antivirulence approach for Gram-negative pathogens
- PMID: 25389178
- PMCID: PMC4235214
- DOI: 10.1128/mBio.02165-14
QseC inhibitors as an antivirulence approach for Gram-negative pathogens
Abstract
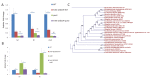
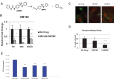
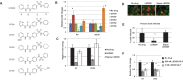
Invasive pathogens interface with the host and its resident microbiota through interkingdom signaling. The bacterial receptor QseC, which is a membrane-bound histidine sensor kinase, responds to the host stress hormones epinephrine and norepinephrine and the bacterial signal AI-3, integrating interkingdom signaling at the biochemical level. Importantly, the QseC signaling cascade is exploited by many bacterial pathogens to promote virulence. Here, we translated this basic science information into development of a potent small molecule inhibitor of QseC, LED209. Extensive structure activity relationship (SAR) studies revealed that LED209 is a potent prodrug that is highly selective for QseC. Its warhead allosterically modifies lysines in QseC, impairing its function and preventing the activation of the virulence program of several Gram-negative pathogens both in vitro and during murine infection. LED209 does not interfere with pathogen growth, possibly leading to a milder evolutionary pressure toward drug resistance. LED209 has desirable pharmacokinetics and does not present toxicity in vitro and in rodents. This is a unique antivirulence approach, with a proven broad-spectrum activity against multiple Gram-negative pathogens that cause mammalian infections.
Importance: There is an imminent need for development of novel treatments for infectious diseases, given that one of the biggest challenges to medicine in the foreseeable future is the emergence of microbial antibiotic resistance. Here, we devised a broad-spectrum antivirulence approach targeting a conserved histidine kinase, QseC, in several Gram-negative pathogens that promotes their virulence expression. The LED209 QseC inhibitor has a unique mode of action by acting as a prodrug scaffold to deliver a warhead that allosterically modifies QseC, impeding virulence in several Gram-negative pathogens.
Copyright © 2014 Curtis et al.
Figures



References
-
- Kostakioti M, Hadjifrangiskou M, Cusumano CK, Hannan TJ, Janetka JW, Hultgren SJ. 2012. Distinguishing the contribution of type 1 pili from that of other QseB-misregulated factors when QseC is absent during urinary tract infection. Infect. Immun. 80:2826–2834. 10.1128/IAI.00283-12. - DOI - PMC - PubMed
-
- Rasko DA, Moreira CG, Li de R, Reading NC, Ritchie JM, Waldor MK, Williams N, Taussig R, Wei S, Roth M, Hughes DT, Huntley JF, Fina MW, Falck JR, Sperandio V. 2008. Targeting QseC signaling and virulence for antibiotic development. Science 321:1078–1080. 10.1126/science.1160354. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
