Characterization and comparative analysis of antibiotic resistance plasmids isolated from a wastewater treatment plant
- PMID: 25389419
- PMCID: PMC4211555
- DOI: 10.3389/fmicb.2014.00558
Characterization and comparative analysis of antibiotic resistance plasmids isolated from a wastewater treatment plant
Abstract
A wastewater treatment plant (WWTP) is an environment high in nutrient concentration with diverse bacterial populations and can provide an ideal environment for the proliferation of mobile elements such as plasmids. WWTPs have also been identified as reservoirs for antibiotic resistance genes that are associated with human pathogens. The objectives of this study were to isolate and characterize self-transmissible or mobilizable resistance plasmids associated with effluent from WWTP. An enrichment culture approach designed to capture plasmids conferring resistance to high concentrations of erythromycin was used to capture plasmids from an urban WWTP servicing a population of ca. 210,000. DNA sequencing of the plasmids revealed diversity of plasmids represented by incompatibility groups IncU, col-E, IncFII and IncP-1β. Genes coding resistance to clinically relevant antibiotics (macrolide, tetracycline, beta-lactam, trimethoprim, chloramphenicol, sulphonamide), quaternary ammonium compounds and heavy metals were co-located on these plasmids, often within transposable and integrative mobile elements. Several of the plasmids were self-transmissible or mobilizable and could be maintained in the absence of antibiotic selection. The IncFII plasmid pEFC36a showed the highest degree of sequence identity to plasmid R1 which has been isolated in England more than 50 years ago from a patient suffering from a Salmonella infection. Functional conservation of key regulatory features of this F-like conjugation module were demonstrated by the finding that the conjugation frequency of pEFC36a could be stimulated by the positive regulator of plasmid R1 DNA transfer genes, TraJ.
Keywords: antibiotic resistance genes; conjugation; genetic; mobile genetic elements; plasmids; wastewater.
Figures
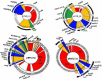
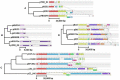
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

