The Cytochrome P450 superfamily complement (CYPome) in the annelid Capitella teleta
- PMID: 25390889
- PMCID: PMC4229089
- DOI: 10.1371/journal.pone.0107728
The Cytochrome P450 superfamily complement (CYPome) in the annelid Capitella teleta
Abstract
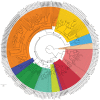
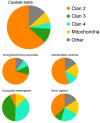
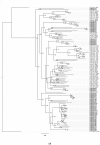
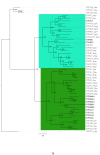
The Cytochrome P450 super family (CYP) is responsible for a wide range of functions in metazoans, having roles in both exogenous and endogenous substrate metabolism. Annelids are known to metabolize polycyclic aromatic hydrocarbons (PAHs) and produce estrogen. CYPs are postulated to be key enzymes in these processes in annelids. In this study, the CYP complement (CYPome) of the annelid Capitella teleta has been robustly identified and annotated with the genome assembly available. Phylogenetic analyses were performed to understand the evolutionary relationships between CYPs in C. teleta and other species. Predictions of which CYPs are potentially involved in both PAH metabolism and steroidogensis were made based on phylogeny. Annotation of 84 full length and 12 partial CYP sequences predicted a total of 96 functional CYPs in C. teleta. A further 13 CYP fragments were found but these may be pseudogenes. The C. teleta CYPome contained 24 novel CYP families and seven novel CYP subfamilies within existing families. A phylogenetic analysis identified that the C. teleta sequences were found in 9 of the 11 metazoan CYP clans. Two CYPs, CYP3071A1 and CYP3072A1, did not cluster with any metazoan CYP clans. We found xenobiotic response elements (XREs) upstream of C. teleta CYPs related to vertebrate CYP1 (CYP3060A1, CYP3061A1) and from families with reported transcriptional upregulation in response to PAH exposure (CYP4, CYP331). C. teleta had a CYP51A1 with ∼65% identity to vertebrate CYP51A1 sequences and has been predicted to have lanosterol 14 α-demethylase activity. CYP376A1, CYP3068A1, CYP3069A1, and CYP3070A1 were the most appropriate candidates for steroidogenesis genes based on their phylogeny and warrant further analyses, though no specific aromatase (estrogen synthesis) candidates were found. Presence of XREs upstream of C. teleta CYPs may indicate a functional aryl hydrocarbon receptor in C. teleta and candidate CYPs for studies of PAH metabolism.
Conflict of interest statement
Figures
References
-
- Nelson DR (2011) Progress in tracing the evolutionary paths of cytochrome P450. BBA Proteins Proteom 1814: 14–18. - PubMed
-
- Nelson DR (1999) Cytochrome P450 and the individuality of species. Arch Biochem Biophys 369: 1–10. - PubMed
-
- Nebert DW, Gonzalez FJ (1987) P450 genes: Structure, evolution, and regulation. Annu Rev Biochem 56: 945–993. - PubMed
-
- Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, et al. (1996) P450 superfamily: Update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenet Genomics 6: 1–42. - PubMed
-
- Lewis DFV (2004) 57 varieties: The human cytochromes P450. Pharmacogenomics 5: 305–318. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

