Conformational dynamics of the focal adhesion targeting domain control specific functions of focal adhesion kinase in cells
- PMID: 25391654
- PMCID: PMC4281750
- DOI: 10.1074/jbc.M114.593632
Conformational dynamics of the focal adhesion targeting domain control specific functions of focal adhesion kinase in cells
Abstract
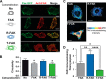
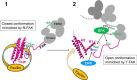
Focal adhesion (FA) kinase (FAK) regulates cell survival and motility by transducing signals from membrane receptors. The C-terminal FA targeting (FAT) domain of FAK fulfils multiple functions, including recruitment to FAs through paxillin binding. Phosphorylation of FAT on Tyr(925) facilitates FA disassembly and connects to the MAPK pathway through Grb2 association, but requires dissociation of the first helix (H1) of the four-helix bundle of FAT. We investigated the importance of H1 opening in cells by comparing the properties of FAK molecules containing wild-type or mutated FAT with impaired or facilitated H1 openings. These mutations did not alter the activation of FAK, but selectively affected its cellular functions, including self-association, Tyr(925) phosphorylation, paxillin binding, and FA targeting and turnover. Phosphorylation of Tyr(861), located between the kinase and FAT domains, was also enhanced by the mutation that opened the FAT bundle. Similarly phosphorylation of Ser(910) by ERK in response to bombesin was increased by FAT opening. Although FAK molecules with the mutation favoring FAT opening were poorly recruited at FAs, they efficiently restored FA turnover and cell shape in FAK-deficient cells. In contrast, the mutation preventing H1 opening markedly impaired FAK function. Our data support the biological importance of conformational dynamics of the FAT domain and its functional interactions with other parts of the molecule.
Keywords: Conformational Change; Focal Adhesions; Protein Kinase; Protein Structure; Tyrosine-Protein Kinase (Tyrosine Kinase).
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
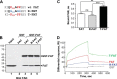
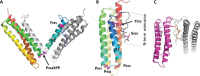


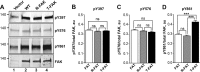

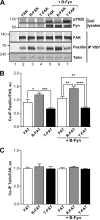

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

