Closing the MCM cycle at replication termination sites
- PMID: 25391904
- PMCID: PMC4264923
- DOI: 10.15252/embr.201439774
Closing the MCM cycle at replication termination sites
Abstract
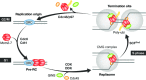
The initiation of eukaryotic DNA replication is a highly regulated process conserved from yeast to human. The past decade has seen significant advances in understanding how the CMG (Cdc45‐MCM‐GINS) replicative helicase is loaded onto DNA. However, very little was known on how this complex is removed from chromatin at the end of S phase. Two papers in a recent issue of Science [1], [2] show that in yeast and in Xenopus, the CMG complex is unloaded at replication termination sites by an active mechanism involving the polyubiquitylation of Mcm7.
Figures
Comment on
-
Polyubiquitylation drives replisome disassembly at the termination of DNA replication.Science. 2014 Oct 24;346(6208):477-81. doi: 10.1126/science.1253585. Science. 2014. PMID: 25342805
-
Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication.Science. 2014 Oct 24;346(6208):1253596. doi: 10.1126/science.1253596. Science. 2014. PMID: 25342810 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

