Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice
- PMID: 25392068
- PMCID: PMC4301742
- DOI: 10.1093/pcp/pcu154
Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice
Abstract
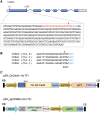
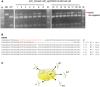
The clustered regularly interspaced short palindromic repeat (CRISPR)-associated endonuclease 9 (CRISPR/Cas9) system has been demonstrated to be a robust genome engineering tool in a variety of organisms including plants. However, it has been shown that the CRISPR/Cas9 system cleaves genomic DNA sequences containing mismatches to the guide RNA strand. We expected that this low specificity could be exploited to induce multihomeologous and multiparalogous gene knockouts. In the case of polyploid plants, simultaneous modification of multiple homeologous genes, i.e. genes with similar but not identical DNA sequences, is often needed to obtain a desired phenotype. Even in diploid plants, disruption of multiparalogous genes, which have functional redundancy, is often needed. To validate the applicability of the CRISPR/Cas9 system to target mutagenesis of paralogous genes in rice, we designed a single-guide RNA (sgRNA) that recognized 20 bp sequences of cyclin-dependent kinase B2 (CDKB2) as an on-target locus. These 20 bp possess similarity to other rice CDK genes (CDKA1, CDKA2 and CDKB1) with different numbers of mismatches. We analyzed mutations in these four CDK genes in plants regenerated from Cas9/sgRNA-transformed calli and revealed that single, double and triple mutants of CDKA2, CDKB1 and CDKB2 can be created by a single sgRNA.
Keywords: CDK; CRISPR/Cas9; Off-target mutation; Rice.
© The Author 2014. Published by Oxford University Press on behalf of Japanese Society of Plant Physiologists.
Figures

References
-
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

