The nature of the N-terminal amino acid residue of HIV-1 RNase H is critical for the stability of reverse transcriptase in viral particles
- PMID: 25392207
- PMCID: PMC4300642
- DOI: 10.1128/JVI.02312-14
The nature of the N-terminal amino acid residue of HIV-1 RNase H is critical for the stability of reverse transcriptase in viral particles
Abstract
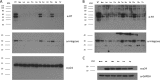
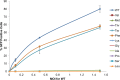
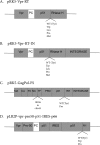
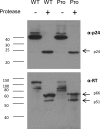
Reverse transcriptase (RT) of human immunodeficiency virus type 1 (HIV-1) is synthesized and packaged into the virion as a part of the GagPol polyprotein. Mature RT is released by the action of viral protease. However, unlike other viral proteins, RT is subject to an internal cleavage event leading to the formation of two subunits in the virion: a p66 subunit and a p51 subunit that lacks the RNase H domain. We have previously identified RNase H to be an HIV-1 protein that has the potential to be a substrate for the N-end rule pathway, which is an ubiquitin-dependent proteolytic system in which the identity of the N-terminal amino acid determines the half-life of a protein. Here we examined the importance of the N-terminal amino acid residue of RNase H in the early life cycle of HIV-1. We show that changing this residue to an amino acid structurally different from the conserved residue leads to the degradation of RT and, in some cases, integrase in the virus particle and this abolishes infectivity. Using intravirion complementation and in vitro protease cleavage assays, we show that degradation of RT in RNase H N-terminal mutants occurs in the absence of active viral protease in the virion. Our results also indicate the importance of the RNase H N-terminal residue in the dimerization of RT subunits.
Importance: HIV-1 proteins are initially made as part of a polyprotein that is cleaved by the viral protease into the proteins that form the virus particle. We were interested in one particular protein, RNase H, that is cleaved from reverse transcriptase. In particular, we found that the first amino acid of RNase H never varied in over 1,850 isolates of HIV-1 that we compared. When we changed the first amino acid, we found that the reverse transcriptase in the virus was degraded. While other studies have implied that the viral protease can degrade mutant RT proteins, we show here that this may not be the case for our mutants. Our results suggest that the presence of active viral protease is not required for the degradation of RT in RNase H N-terminal mutants, suggesting a role for a cellular protease in this process.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures






Similar articles
-
The ribonuclease H activity of the reverse transcriptases of human immunodeficiency viruses type 1 and type 2 is affected by the thumb subdomain of the small protein subunits.J Mol Biol. 2001 Aug 31;311(5):957-71. doi: 10.1006/jmbi.2001.4904. J Mol Biol. 2001. PMID: 11531332
-
Virion instability of human immunodeficiency virus type 1 reverse transcriptase (RT) mutated in the protease cleavage site between RT p51 and the RT RNase H domain.J Virol. 2005 Sep;79(18):11952-61. doi: 10.1128/JVI.79.18.11952-11961.2005. J Virol. 2005. PMID: 16140771 Free PMC article.
-
Mutagenesis of cysteine 280 of the reverse transcriptase of human immunodeficiency virus type-1: the effects on the ribonuclease H activity.J Mol Biol. 2003 Mar 14;327(1):19-30. doi: 10.1016/s0022-2836(03)00052-4. J Mol Biol. 2003. PMID: 12614605
-
Human Immunodeficiency Virus type 1 reverse transcriptase.Biol Chem Hoppe Seyler. 1996 Feb;377(2):97-120. Biol Chem Hoppe Seyler. 1996. PMID: 8868066 Review.
-
Search for new therapeutics against HIV-1 via dual inhibition of RNase H and integrase: current status and future challenges.Future Med Chem. 2021 Feb;13(3):269-286. doi: 10.4155/fmc-2020-0257. Epub 2021 Jan 5. Future Med Chem. 2021. PMID: 33399497 Review.
Cited by
-
Structural integrity of the ribonuclease H domain in HIV-1 reverse transcriptase.Proteins. 2015 Aug;83(8):1526-38. doi: 10.1002/prot.24843. Epub 2015 Jul 1. Proteins. 2015. PMID: 26061827 Free PMC article.
-
Molecular Docking Studies of HIV-1 Resistance to Reverse Transcriptase Inhibitors: Mini-Review.Molecules. 2018 May 21;23(5):1233. doi: 10.3390/molecules23051233. Molecules. 2018. PMID: 29883406 Free PMC article. Review.
-
Isolation of amaranthin synthetase from Chenopodium quinoa and construction of an amaranthin production system using suspension-cultured tobacco BY-2 cells.Plant Biotechnol J. 2019 May;17(5):969-981. doi: 10.1111/pbi.13032. Epub 2018 Dec 5. Plant Biotechnol J. 2019. PMID: 30451369 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

