Biological and protective properties of immune sera directed to the influenza virus neuraminidase
- PMID: 25392225
- PMCID: PMC4300740
- DOI: 10.1128/JVI.02949-14
Biological and protective properties of immune sera directed to the influenza virus neuraminidase
Abstract
The envelope of influenza A viruses contains two large antigens, hemagglutinin (HA) and neuraminidase (NA). Conventional influenza virus vaccines induce neutralizing antibodies that are predominantly directed to the HA globular head, a domain that is subject to extensive antigenic drift. Antibodies directed to NA are induced at much lower levels, probably as a consequence of the immunodominance of the HA antigen. Although antibodies to NA may affect virus release by inhibiting the sialidase function of the glycoprotein, the antigen has been largely neglected in past vaccine design. In this study, we characterized the protective properties of monospecific immune sera that were generated by vaccination with recombinant RNA replicon particles encoding NA. These immune sera inhibited hemagglutination in an NA subtype-specific and HA subtype-independent manner and interfered with infection of MDCK cells. In addition, they inhibited the sialidase activities of various influenza viruses of the same and even different NA subtypes. With this, the anti-NA immune sera inhibited the spread of H5N1 highly pathogenic avian influenza virus and HA/NA-pseudotyped viruses in MDCK cells in a concentration-dependent manner. When chickens were immunized with NA recombinant replicon particles and subsequently infected with low-pathogenic avian influenza virus, inflammatory serum markers were significantly reduced and virus shedding was limited or eliminated. These findings suggest that NA antibodies can inhibit virus dissemination by interfering with both virus attachment and egress. Our results underline the potential of high-quality NA antibodies for controlling influenza virus replication and place emphasis on NA as a vaccine antigen.
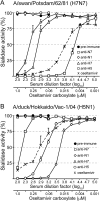
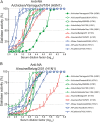
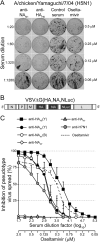
Importance: The neuraminidase of influenza A viruses is a sialidase that acts as a receptor-destroying enzyme facilitating the release of progeny virus from infected cells. Here, we demonstrate that monospecific anti-NA immune sera inhibited not only sialidase activity, but also influenza virus hemagglutination and infection of MDCK cells, suggesting that NA antibodies can interfere with virus attachment. Inhibition of both processes, virus release and virus binding, may explain why NA antibodies efficiently blocked virus dissemination in vitro and in vivo. Anti-NA immune sera showed broader reactivity than anti-HA sera in hemagglutination inhibition tests and demonstrated cross-subtype activity in sialidase inhibition tests. These remarkable features of NA antibodies highlight the importance of the NA antigen for the development of next-generation influenza virus vaccines.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




References
-
- Lotan R, Skutelsky E, Danon D, Sharon N. 1975. The purification, composition, and specificity of the anti-T lectin from peanut (Arachis hypogaea). J Biol Chem 250:8518–8523. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

