Genomic signature of multidrug-resistant Salmonella enterica serovar typhi isolates related to a massive outbreak in Zambia between 2010 and 2012
- PMID: 25392358
- PMCID: PMC4290967
- DOI: 10.1128/JCM.02026-14
Genomic signature of multidrug-resistant Salmonella enterica serovar typhi isolates related to a massive outbreak in Zambia between 2010 and 2012
Abstract
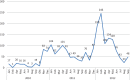
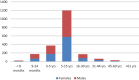
Retrospectively, we investigated the epidemiology of a massive Salmonella enterica serovar Typhi outbreak in Zambia during 2010 to 2012. Ninety-four isolates were susceptibility tested by MIC determinations. Whole-genome sequence typing (WGST) of 33 isolates and bioinformatic analysis identified the multilocus sequence type (MLST), haplotype, plasmid replicon, antimicrobial resistance genes, and genetic relatedness by single nucleotide polymorphism (SNP) analysis and genomic deletions. The outbreak affected 2,040 patients, with a fatality rate of 0.5%. Most (83.0%) isolates were multidrug resistant (MDR). The isolates belonged to MLST ST1 and a new variant of the haplotype, H58B. Most isolates contained a chromosomally translocated region containing seven antimicrobial resistance genes, catA1, blaTEM-1, dfrA7, sul1, sul2, strA, and strB, and fragments of the incompatibility group Q1 (IncQ1) plasmid replicon, the class 1 integron, and the mer operon. The genomic analysis revealed 415 SNP differences overall and 35 deletions among 33 of the isolates subjected to whole-genome sequencing. In comparison with other genomes of H58, the Zambian isolates separated from genomes from Central Africa and India by 34 and 52 SNPs, respectively. The phylogenetic analysis indicates that 32 of the 33 isolates sequenced belonged to a tight clonal group distinct from other H58 genomes included in the study. The small numbers of SNPs identified within this group are consistent with the short-term transmission that can be expected over a period of 2 years. The phylogenetic analysis and deletions suggest that a single MDR clone was responsible for the outbreak, during which occasional other S. Typhi lineages, including sensitive ones, continued to cocirculate. The common view is that the emerging global S. Typhi haplotype, H58B, containing the MDR IncHI1 plasmid is responsible for the majority of typhoid infections in Asia and sub-Saharan Africa; we found that a new variant of the haplotype harboring a chromosomally translocated region containing the MDR islands of IncHI1 plasmid has emerged in Zambia. This could change the perception of the term "classical MDR typhoid" currently being solely associated with the IncHI1 plasmid. It might be more common than presently thought that S. Typhi haplotype H58B harbors the IncHI1 plasmid or a chromosomally translocated MDR region or both.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



References
-
- Hendriksen RS, Vieira AR, Karlsmose S, Lo Fo Wong DM, Jensen AB, Wegener HC, Aarestrup FM. 14 April 2011, posting date Global monitoring of Salmonella serovar distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog Dis doi:10.1089/fpd.2010.0787. - DOI - PubMed
-
- Mogasale V, Maskery B, Ochiai RL, Lee JS, Mogasale VV, Ramani E, Kim YE, Park JK, Wierzba TF. 2014. Burden of typhoid fever in low-income and middle-income countries: a systematic, literature-based update with risk-factor adjustment. Lancet Glob Health 2:e570–e580. doi:10.1016/S2214-109X(14)70301-8. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

