Neurogenin3 restricts serotonergic neuron differentiation to the hindbrain
- PMID: 25392491
- PMCID: PMC6608453
- DOI: 10.1523/JNEUROSCI.3403-14.2014
Neurogenin3 restricts serotonergic neuron differentiation to the hindbrain
Erratum in
- J Neurosci. 2015 May 20;35(20):8037
Abstract
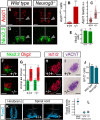
The development of the nervous system is critically dependent on the production of functionally diverse neuronal cell types at their correct locations. In the embryonic neural tube, dorsoventral signaling has emerged as a fundamental mechanism for generating neuronal diversity. In contrast, far less is known about how different neuronal cell types are organized along the rostrocaudal axis. In the developing mouse and chick neural tube, hindbrain serotonergic neurons and spinal glutamatergic V3 interneurons are produced from ventral p3 progenitors, which possess a common transcriptional identity but are confined to distinct anterior-posterior territories. In this study, we show that the expression of the transcription factor Neurogenin3 (Neurog3) in the spinal cord controls the correct specification of p3-derived neurons. Gain- and loss-of-function manipulations in the chick and mouse embryo show that Neurog3 switches ventral progenitors from a serotonergic to V3 differentiation program by repressing Ascl1 in spinal p3 progenitors through a mechanism dependent on Hes proteins. In this way, Neurog3 establishes the posterior boundary of the serotonergic system by actively suppressing serotonergic specification in the spinal cord. These results explain how equivalent p3 progenitors within the hindbrain and the spinal cord produce functionally distinct neuron cell types.
Keywords: hindbrain; neural tube; neuronal specification; serotonergic system; spinal cord; transcription factor.
Copyright © 2014 the authors 0270-6474/14/3415223-11$15.00/0.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
