Clonal architectures and driver mutations in metastatic melanomas
- PMID: 25393105
- PMCID: PMC4230926
- DOI: 10.1371/journal.pone.0111153
Clonal architectures and driver mutations in metastatic melanomas
Abstract
To reveal the clonal architecture of melanoma and associated driver mutations, whole genome sequencing (WGS) and targeted extension sequencing were used to characterize 124 melanoma cases. Significantly mutated gene analysis using 13 WGS cases and 15 additional paired extension cases identified known melanoma genes such as BRAF, NRAS, and CDKN2A, as well as a novel gene EPHA3, previously implicated in other cancer types. Extension studies using tumors from another 96 patients discovered a large number of truncation mutations in tumor suppressors (TP53 and RB1), protein phosphatases (e.g., PTEN, PTPRB, PTPRD, and PTPRT), as well as chromatin remodeling genes (e.g., ASXL3, MLL2, and ARID2). Deep sequencing of mutations revealed subclones in the majority of metastatic tumors from 13 WGS cases. Validated mutations from 12 out of 13 WGS patients exhibited a predominant UV signature characterized by a high frequency of C->T transitions occurring at the 3' base of dipyrimidine sequences while one patient (MEL9) with a hypermutator phenotype lacked this signature. Strikingly, a subclonal mutation signature analysis revealed that the founding clone in MEL9 exhibited UV signature but the secondary clone did not, suggesting different mutational mechanisms for two clonal populations from the same tumor. Further analysis of four metastases from different geographic locations in 2 melanoma cases revealed phylogenetic relationships and highlighted the genetic alterations responsible for differential drug resistance among metastatic tumors. Our study suggests that clonal evaluation is crucial for understanding tumor etiology and drug resistance in melanoma.
Conflict of interest statement
Figures


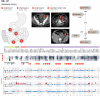
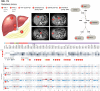
Similar articles
-
[Characterization of genetic alterations in primary human melanomas carrying BRAF or NRAS mutation].Magy Onkol. 2013 Jun;57(2):96-9. Epub 2013 May 20. Magy Onkol. 2013. PMID: 23795354 Hungarian.
-
Clinicopathological characteristics and mutation profiling in primary cutaneous melanoma.Am J Dermatopathol. 2015 May;37(5):389-97. doi: 10.1097/DAD.0000000000000241. Am J Dermatopathol. 2015. PMID: 25357015
-
[ Spectrum of oncogene mutations is different in melanoma subtypes].Mol Biol (Mosk). 2015 Nov-Dec;49(6):1022-9. doi: 10.7868/S0026898415060166. Mol Biol (Mosk). 2015. PMID: 26710785 Russian.
-
Mutation stability in primary and metastatic melanoma: what we know and what we don't.Histol Histopathol. 2015 Jul;30(7):763-70. doi: 10.14670/HH-11-584. Epub 2015 Jan 13. Histol Histopathol. 2015. PMID: 25585249 Review.
-
Somatic driver mutations in melanoma.Cancer. 2017 Jun 1;123(S11):2104-2117. doi: 10.1002/cncr.30593. Cancer. 2017. PMID: 28543693 Review.
Cited by
-
Cutaneous Melanoma Classification: The Importance of High-Throughput Genomic Technologies.Front Oncol. 2021 May 28;11:635488. doi: 10.3389/fonc.2021.635488. eCollection 2021. Front Oncol. 2021. PMID: 34123788 Free PMC article. Review.
-
From fish bowl to bedside: The power of zebrafish to unravel melanoma pathogenesis and discover new therapeutics.Pigment Cell Melanoma Res. 2017 Jul;30(4):402-412. doi: 10.1111/pcmr.12592. Epub 2017 Jun 8. Pigment Cell Melanoma Res. 2017. PMID: 28379616 Free PMC article. Review.
-
Evolution of delayed resistance to immunotherapy in a melanoma responder.Nat Med. 2021 Jun;27(6):985-992. doi: 10.1038/s41591-021-01331-8. Epub 2021 May 3. Nat Med. 2021. PMID: 33941922 Free PMC article.
-
Nanoparticle-Based Combination Therapy for Melanoma.Front Oncol. 2022 Jun 28;12:928797. doi: 10.3389/fonc.2022.928797. eCollection 2022. Front Oncol. 2022. PMID: 35837089 Free PMC article. Review.
-
Late-Stage Metastatic Melanoma Emerges through a Diversity of Evolutionary Pathways.Cancer Discov. 2023 Jun 2;13(6):1364-1385. doi: 10.1158/2159-8290.CD-22-1427. Cancer Discov. 2023. PMID: 36977461 Free PMC article.
References
-
- Siegel R, Naishadham D, Jemal A (2013) Cancer statistics, 2013. CA: a cancer journal for clinicians 63: 11–30. - PubMed
-
- Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, et al. (2005) Distinct sets of genetic alterations in melanoma. The New England journal of medicine 353: 2135–2147. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

