Cyclooxygenase-2 silencing for the treatment of colitis: a combined in vivo strategy based on RNA interference and engineered Escherichia coli
- PMID: 25393372
- PMCID: PMC4445621
- DOI: 10.1038/mt.2014.222
Cyclooxygenase-2 silencing for the treatment of colitis: a combined in vivo strategy based on RNA interference and engineered Escherichia coli
Abstract
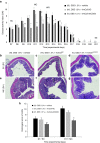
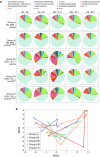
Nonpathogenic-invasive Escherichia coli (InvColi) bacteria are suitable for genetic transfer into mammalian cells and may act as a vehicle for RNA Interference (RNAi) in vivo. Cyclooxygenase-2 (COX-2) is overexpressed in ulcerative colitis (UC) and Crohn's disease (CD), two inflammatory conditions of the colon and small intestine grouped as inflammatory bowel disease (IBD). We engineered InvColi strains for anti-COX-2 RNAi (InvColi(shCOX2)), aiming to investigate the in vivo feasibility of a novel COX-2 silencing strategy in a murine model of colitis induced by dextran sulfate sodium (DSS). Enema administrations of InvColi(shCOX2) in DSS-treated mice led to COX-2 downregulation, colonic mucosa preservation, reduced colitis disease activity index (DAI) and increased mice survival. Moreover, DSS/InvColi(shCOX2)-treated mice showed lower levels of circulating pro-inflammatory cytokines and a reduced colitis-associated shift of gut microbiota. Considering its effectiveness and safety, we propose our InvColi(shCOX2) strategy as a promising tool for molecular therapy in intestinal inflammatory diseases.
Figures



References
-
- Grillot-Courvalin C, Goussard S, Huetz F, Ojcius DM, Courvalin P. Functional gene transfer from intracellular bacteria to mammalian cells. Nat Biotechnol. 1998;16:862–866. - PubMed
-
- Critchley-Thorne RJ, Stagg AJ, Vassaux G. Recombinant Escherichia coli expressing invasin targets the Peyer's patches: the basis for a bacterial formulation for oral vaccination. Mol Ther. 2006;14:183–191. - PubMed
-
- Xiang S, Fruehauf J, Li CJ. Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nat Biotechnol. 2006;24:697–702. - PubMed
-
- Spisni E, Tomasi V. Bicknell R, Lewis CE, Ferrara N. Tumour Angiogenesis. Oxford University Press; Oxford; 1997. Involvement of prostanoid in angiogenesis; pp. pp. 291–300.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

