Quantitative proteomic analysis of serum from pregnant women carrying a fetus with conotruncal heart defect using isobaric tags for relative and absolute quantitation (iTRAQ) labeling
- PMID: 25393621
- PMCID: PMC4230941
- DOI: 10.1371/journal.pone.0111645
Quantitative proteomic analysis of serum from pregnant women carrying a fetus with conotruncal heart defect using isobaric tags for relative and absolute quantitation (iTRAQ) labeling
Abstract
Objective: To identify differentially expressed proteins from serum of pregnant women carrying a conotruncal heart defects (CTD) fetus, using proteomic analysis.
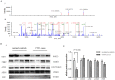
Methods: The study was conducted using a nested case-control design. The 5473 maternal serum samples were collected at 14-18 weeks of gestation. The serum from 9 pregnant women carrying a CTD fetus, 10 with another CHD (ACHD) fetus, and 11 with a normal fetus were selected from the above samples, and analyzed by using isobaric tags for relative and absolute quantitation (iTRAQ) coupled with two-dimensional liquid chromatography-tandem mass spectrometry(2D LC-MS/MS). The differentially expressed proteins identified by iTRAQ were further validated with Western blot.
Results: A total of 105 unique proteins present in the three groups were identified, and relative expression data were obtained for 92 of them with high confidence by employing the iTRAQ-based experiments. The downregulation of gelsolin in maternal serum of fetus with CTD was further verified by Western blot.
Conclusions: The identification of differentially expressed protein gelsolin in the serum of the pregnant women carrying a CTD fetus by using proteomic technology may be able to serve as a foundation to further explore the biomarker for detection of CTD fetus from the maternal serum.
Conflict of interest statement
Figures


References
-
- Botto LD, Correa A, Erickson JD (2001) Racial and temporal variations in the prevalence of heart defects. Pediatrics 107: E32. - PubMed
-
- Centers for Disease Control and Prevention (CDC) (1998) Trends in infant mortality attributable to birth defects—United States, 1980–1995. MMWR Morb Mortal Wkly Rep 47: 773–778. - PubMed
-
- Benson DW, Basson CT, MacRae CA (1996) New understandings in the genetics of congenital heart disease. Curr Opin Pediatr 8: 505–511. - PubMed
-
- Pierpont ME, Basson CT, Benson DW Jr, Gelb BD, Giglia TM, et al. (2007) Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation 115: 3015–3038. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

