Multiple dimensions of epigenetic gene regulation in the malaria parasite Plasmodium falciparum: gene regulation via histone modifications, nucleosome positioning and nuclear architecture in P. falciparum
- PMID: 25394267
- PMCID: PMC4385791
- DOI: 10.1002/bies.201400145
Multiple dimensions of epigenetic gene regulation in the malaria parasite Plasmodium falciparum: gene regulation via histone modifications, nucleosome positioning and nuclear architecture in P. falciparum
Abstract
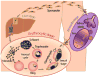
Plasmodium falciparum is the most deadly human malarial parasite, responsible for an estimated 207 million cases of disease and 627,000 deaths in 2012. Recent studies reveal that the parasite actively regulates a large fraction of its genes throughout its replicative cycle inside human red blood cells and that epigenetics plays an important role in this precise gene regulation. Here, we discuss recent advances in our understanding of three aspects of epigenetic regulation in P. falciparum: changes in histone modifications, nucleosome occupancy and the three-dimensional genome structure. We compare these three aspects of the P. falciparum epigenome to those of other eukaryotes, and show that large-scale compartmentalization is particularly important in determining histone decomposition and gene regulation in P. falciparum. We conclude by presenting a gene regulation model for P. falciparum that combines the described epigenetic factors, and by discussing the implications of this model for the future of malaria research.
Keywords: epigenetics; gene regulation; histone modifications; malaria; nucleosome occupancy; three-dimensional genome organization; virulence genes.
© 2015 WILEY Periodicals, Inc.
Conflict of interest statement
The authors have declared no conflict of interest.
Figures


References
-
- Baruch DI, Pasloske BL, Singh HB, Bi X, et al. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. - PubMed
-
- Su XZ, Heatwole VM, Wertheimer SP, Guinet F, et al. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

