MiR-183/-96/-182 cluster is up-regulated in most breast cancers and increases cell proliferation and migration
- PMID: 25394902
- PMCID: PMC4303194
- DOI: 10.1186/s13058-014-0473-z
MiR-183/-96/-182 cluster is up-regulated in most breast cancers and increases cell proliferation and migration
Abstract
Introduction: The miR-183/-96/-182 cluster is a conserved polycistronic microRNA (miRNA) cluster which is highly expressed in most breast cancers. Although there are some sporadic reports which demonstrate the importance of each miRNA in this cluster in breast cancer, the biological roles of this cluster as a whole and its regulation mechanisms in breast cancer are still unclear. We compared the expression of this cluster in different cancer types, analyzed the regulation mechanism of this cluster, identified new target genes, and examined the impact of this cluster on breast cancer cells.
Methods: The miRNA level was detected by LNA-based northern blot and Real-time PCR, and was also analyzed from TCGA dataset. Bioinformatics research and luciferase assay were applied to find the promoter regions and transcription factors. To investigate the biological effects of the miR-183/-96 /-182 cluster in breast cancer, we generated miR-96, miR-182 and miR-183 overexpression stable cell lines to check the overdose effects; we also used miR-Down™ antagomir for each miRNA as well as miR-183/-96 /-182 cluster sponge lentivirus to check the knockdown effects. Growth, migration, cell cycle profile and survival of these cells was then monitored by colony formation assay, MTT assay, cell wound healing assay, flow cytometry and microscopy. The target gene was validated by Real-time PCR, luciferase assay, Western blot and Phalloidin/DAPI counterstaining.
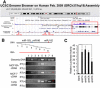
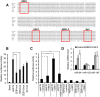
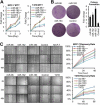
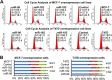
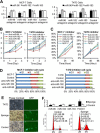
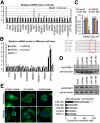
Results: The miR-183/-96/-182 cluster was highly expressed in most breast cancers, and its transcription is disordered in breast cancer. The miR-183/-96/-182 cluster was transcribed in the same pri-miRNA and its transcription was regulated by ZEB1 and HSF2. It increased breast cell growth by promoting more rapid completion of mitosis, promoted cell migration and was essential for cell survival. MiR-183 targeted the RAB21 mRNA directly in breast cancer.
Conclusion: The miR-183/-96/-182 cluster is up-regulated in most breast cancer. It functions as an oncogene in breast cancer as it increases cell proliferation and migration.
Figures

References
-
- Xu D, He X, Chang Y, Xu C, Jiang X, Sun S, Lin J. Inhibition of miR-96 expression reduces cell proliferation and clonogenicity of HepG2 hepatoma cells. Oncol Rep. 2013;29:653–661. - PubMed
-
- Mihelich BL, Khramtsova EA, Arva N, Vaishnav A, Johnson DN, Giangreco AA, Martens-Uzunova E, Baqasra O, Kajdacsy-Balla A, Nonn L. miR-183–96–182 cluster is overexpressed in prostate tissue and regulates zinc homeostasis in prostate cells. J Biol Chem. 2011;286:44503–44511. doi: 10.1074/jbc.M111.262915. - DOI - PMC - PubMed
-
- Weeraratne SD, Amani V, Teider N, Pierre-Francois J, Winter D, Kye MJ, Senqupta S, Archer T, Remke M, Bai AH, Warren P, Pfister SM, Steen JA, Pomeroy SJ, Cho YJ. Pleiotropic effects of miR-183 ~ 96 ~ 182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol. 2012;123:539–552. doi: 10.1007/s00401-012-0969-5. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

