Pdsg1 and Pdsg2, novel proteins involved in developmental genome remodelling in Paramecium
- PMID: 25397898
- PMCID: PMC4232520
- DOI: 10.1371/journal.pone.0112899
Pdsg1 and Pdsg2, novel proteins involved in developmental genome remodelling in Paramecium
Erratum in
-
Correction: Pdsg1 and Pdsg2, novel proteins involved in developmental genome remodelling in Paramecium.PLoS One. 2015 Feb 25;10(2):e0118384. doi: 10.1371/journal.pone.0118384. eCollection 2015. PLoS One. 2015. PMID: 25714097 Free PMC article. No abstract available.
Abstract
The epigenetic influence of maternal cells on the development of their progeny has long been studied in various eukaryotes. Multicellular organisms usually provide their zygotes not only with nutrients but also with functional elements required for proper development, such as coding and non-coding RNAs. These maternally deposited RNAs exhibit a variety of functions, from regulating gene expression to assuring genome integrity. In ciliates, such as Paramecium these RNAs participate in the programming of large-scale genome reorganization during development, distinguishing germline-limited DNA, which is excised, from somatic-destined DNA. Only a handful of proteins playing roles in this process have been identified so far, including typical RNAi-derived factors such as Dicer-like and Piwi proteins. Here we report and characterize two novel proteins, Pdsg1 and Pdsg2 (Paramecium protein involved in Development of the Somatic Genome 1 and 2), involved in Paramecium genome reorganization. We show that these proteins are necessary for the excision of germline-limited DNA during development and the survival of sexual progeny. Knockdown of PDSG1 and PDSG2 genes affects the populations of small RNAs known to be involved in the programming of DNA elimination (scanRNAs and iesRNAs) and chromatin modification patterns during development. Our results suggest an association between RNA-mediated trans-generational epigenetic signal and chromatin modifications in the process of Paramecium genome reorganization.
Conflict of interest statement
Figures

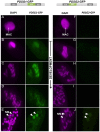
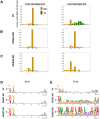

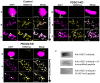
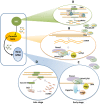
References
-
- Betermier M (2004) Large-scale genome remodelling by the developmentally programmed elimination of germ line sequences in the ciliate Paramecium. Res Microbiol 155: 399–408. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

