Applications of hydrogen/deuterium exchange MS from 2012 to 2014
- PMID: 25398026
- PMCID: PMC4287169
- DOI: 10.1021/ac5040242
Applications of hydrogen/deuterium exchange MS from 2012 to 2014
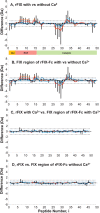
Figures









References
-
- Jankowska E.; Stefanowicz P.; Sosnowska M.; Karpowicz P.; Radziszewska K.; Szewczuk Z.; Szymanska A. Proteins 2012, 80, 2417–2425. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

