Carcinogenic polycyclic aromatic hydrocarbons induce CYP1A1 in human cells via a p53-dependent mechanism
- PMID: 25398514
- PMCID: PMC4748000
- DOI: 10.1007/s00204-014-1409-1
Carcinogenic polycyclic aromatic hydrocarbons induce CYP1A1 in human cells via a p53-dependent mechanism
Abstract
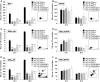
The tumour suppressor gene TP53 is mutated in more than 50 % of human tumours, making it one of the most important cancer genes. We have investigated the role of TP53 in cytochrome P450 (CYP)-mediated metabolic activation of three polycyclic aromatic hydrocarbons (PAHs) in a panel of isogenic colorectal HCT116 cells with differing TP53 status. Cells that were TP53(+/+), TP53(+/-), TP53(-/-), TP53(R248W/+) or TP53(R248W/-) were treated with benzo[a]pyrene (BaP), dibenz[a,h]anthracene and dibenzo[a,l]pyrene, and the formation of DNA adducts was measured by (32)P-postlabelling analysis. Each PAH formed significantly higher DNA adduct levels in TP53(+/+) cells than in the other cell lines. There were also significantly lower levels of PAH metabolites in the culture media of these other cell lines. Bypass of the need for metabolic activation by treating cells with the corresponding reactive PAH-diol-epoxide metabolites resulted in similar adduct levels in all cell lines, which confirms that the influence of p53 is on the metabolism of the parent PAHs. Western blotting showed that CYP1A1 protein expression was induced to much greater extent in TP53(+/+) cells than in the other cell lines. CYP1A1 is inducible via the aryl hydrocarbon receptor (AHR), but we did not find that expression of AHR was dependent on p53; rather, we found that BaP-induced CYP1A1 expression was regulated through p53 binding to a p53 response element in the CYP1A1 promoter region, thereby enhancing its transcription. This study demonstrates a new pathway for CYP1A1 induction by environmental PAHs and reveals an emerging role for p53 in xenobiotic metabolism.
Keywords: Benzo[a]pyrene; Carcinogen metabolism; Cytochrome P450; DNA adducts; Tumour suppressor p53.
Figures




References
-
- Arlt VM, Stiborova M, Henderson CJ, Osborne MR, Bieler CA, Frei E, Martinek V, Sopko B, Wolf CR, Schmeiser HH, Phillips DH. Environmental pollutant and potent mutagen 3-nitrobenzanthrone forms DNA adducts after reduction by NAD(P)H:quinone oxidoreductase and conjugation by acetyltransferases and sulfotransferases in human hepatic cytosols. Cancer Res. 2005;65:2644–2652. doi: 10.1158/0008-5472.CAN-04-3544. - DOI - PubMed
-
- Arlt VM, Schmeiser HH, Osborne MR, et al. Identification of three major DNA adducts formed by the carcinogenic air pollutant 3-nitrobenzanthrone in rat lung at the C8 and N2 position of guanine and at the N6 position of adenine. Int J Cancer. 2006;118:2139–2146. doi: 10.1002/ijc.21622. - DOI - PubMed
-
- Arlt VM, Stiborova M, vom Brocke J, Simoes ML, Lord GM, Nortier JL, Hollstein M, Phillips DH, Schmeiser HH. Aristolochic acid mutagenesis: molecular clues to the aetiology of Balkan endemic nephropathy-associated urothelial cancer. Carcinogenesis. 2007;28:2253–2261. doi: 10.1093/carcin/bgm082. - DOI - PubMed
-
- Arlt VM, Stiborova M, Henderson CJ, Thiemann M, Frei E, Aimova D, Singh R, Gamboa da Costa G, Schmitz OJ, Farmer PB, Wolf CR, Phillips DH. Metabolic activation of benzo[a]pyrene in vitro by hepatic cytochrome P450 contrasts with detoxification in vivo: experiments with hepatic cytochrome P450 reductase null mice. Carcinogenesis. 2008;29:656–665. doi: 10.1093/carcin/bgn002. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

