Ecological succession and viability of human-associated microbiota on restroom surfaces
- PMID: 25398865
- PMCID: PMC4277569
- DOI: 10.1128/AEM.03117-14
Ecological succession and viability of human-associated microbiota on restroom surfaces
Abstract
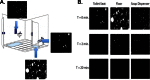
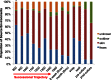
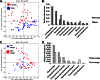
Human-associated bacteria dominate the built environment (BE). Following decontamination of floors, toilet seats, and soap dispensers in four public restrooms, in situ bacterial communities were characterized hourly, daily, and weekly to determine their successional ecology. The viability of cultivable bacteria, following the removal of dispersal agents (humans), was also assessed hourly. A late-successional community developed within 5 to 8 h on restroom floors and showed remarkable stability over weeks to months. Despite late-successional dominance by skin- and outdoor-associated bacteria, the most ubiquitous organisms were predominantly gut-associated taxa, which persisted following exclusion of humans. Staphylococcus represented the majority of the cultivable community, even after several hours of human exclusion. Methicillin-resistant Staphylococcus aureus (MRSA)-associated virulence genes were found on floors but were not present in assembled Staphylococcus pan-genomes. Viral abundances, which were predominantly enterophages, human papilloma virus, and herpesviruses, were significantly correlated with bacterial abundances and showed an unexpectedly low virus-to-bacterium ratio in surface-associated samples, suggesting that bacterial hosts are mostly dormant on BE surfaces.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



References
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

