CNV instability associated with DNA replication dynamics: evidence for replicative mechanisms in CNV mutagenesis
- PMID: 25398944
- PMCID: PMC4381758
- DOI: 10.1093/hmg/ddu572
CNV instability associated with DNA replication dynamics: evidence for replicative mechanisms in CNV mutagenesis
Abstract
Copy number variation (CNV) in the human genome is of vital importance to human health and evolution of our species. However, much of the molecular basis of CNV mutagenesis remains to be elucidated. Considering the DNA replication model of 'fork stalling and template switching' for CNV formation, we hypothesized that replication fork progression could be important for CNV mutagenesis. However, molecular assays of replication fork progression at the genome level are technically challenging. Instead, we conducted an estimation of DNA replication dynamics, as the statistic R, using the readily available data of replication timing. Small R-values can reflect 'slowed' replication, which could result from less fork initiation, reduced fork speed or fork barriers. We generated genome-wide profiles of R in the genomes of human, mouse and Drosophila. Intriguingly, the CNV breakpoints in all three genomes showed significantly biased distributions toward the genomic regions with small R-values, suggesting potential replication stress-induced CNV instability. Notably, among the human CNVs with distinct breakpoint junction characteristics, the homology-mediated and VNTR-mediated CNVs contribute the most to the correlation between CNV instability and the statistic R, consistent with the recent findings in the C. elegans and yeast genomes of repeat-induced DNA replication error and consequent CNV formation. The statistic R may reflect both replication stress and the effect of local genome architecture on fork progression. Our concordant observations suggest an important role for DNA replicative mechanisms in CNV mutagenesis and genome instability.
© The Author 2014. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures
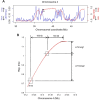
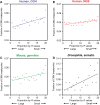
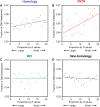
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

