A network biology workflow to study transcriptomics data of the diabetic liver
- PMID: 25399255
- PMCID: PMC4246458
- DOI: 10.1186/1471-2164-15-971
A network biology workflow to study transcriptomics data of the diabetic liver
Abstract
Background: Nowadays a broad collection of transcriptomics data is publicly available in online repositories. Methods for analyzing these data often aim at deciphering the influence of gene expression at the process level. Biological pathway diagrams depict known processes and capture the interactions of gene products and metabolites, information that is essential for the computational analysis and interpretation of transcriptomics data.The present study describes a comprehensive network biology workflow that integrates differential gene expression in the human diabetic liver with pathway information by building a network of interconnected pathways. Worldwide, the incidence of type 2 diabetes mellitus is increasing dramatically, and to better understand this multifactorial disease, more insight into the concerted action of the disease-related processes is needed. The liver is a key player in metabolic diseases and diabetic patients often develop non-alcoholic fatty liver disease.
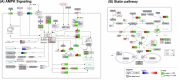
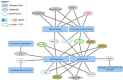
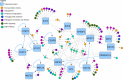
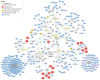
Results: A publicly available dataset comparing the liver transcriptome from lean and healthy vs. obese and insulin-resistant subjects was selected after a thorough analysis. Pathway analysis revealed seven significantly altered pathways in the WikiPathways human pathway collection. These pathways were then merged into one combined network with 408 gene products, 38 metabolites and 5 pathway nodes. Further analysis highlighted 17 nodes present in multiple pathways, and revealed the connections between different pathways in the network. The integration of transcription factor-gene interactions from the ENCODE project identified new links between the pathways on a regulatory level. The extension of the network with known drug-target interactions from DrugBank allows for a more complete study of drug actions and helps with the identification of other drugs that target proteins up- or downstream which might interfere with the action or efficiency of a drug.
Conclusions: The described network biology workflow uses state-of-the-art pathway and network analysis methods to study the rewiring of the diabetic liver. The integration of experimental data and knowledge on disease-affected biological pathways, including regulatory elements like transcription factors or drugs, leads to improved insights and a clearer illustration of the overall process. It also provides a resource for building new hypotheses for further follow-up studies. The approach is highly generic and can be applied in different research fields.
Figures

References
-
- Iozzo P, Hallsten K, Oikonen V, Virtanen KA, Kemppainen J, Solin O, Ferrannini E, Knuuti J, Nuutila P. Insulin-mediated hepatic glucose uptake is impaired in type 2 diabetes: evidence for a relationship with glycemic control. J Clin Endocrinol Metab. 2003;88(5):2055–2060. doi: 10.1210/jc.2002-021446. - DOI - PubMed
-
- Kohjima M, Enjoji M, Higuchi N, Kato M, Kotoh K, Yoshimoto T, Fujino T, Yada M, Yada R, Harada N, Takayanagi R, Nakamuta M. Re-evaluation of fatty acid metabolism-related gene expression in nonalcoholic fatty liver disease. Int J Mol Med. 2007;20(3):351–358. - PubMed
-
- Misu H, Takamura T, Matsuzawa N, Shimizu A, Ota T, Sakurai M, Ando H, Arai K, Yamashita T, Honda M, Kaneko S. Genes involved in oxidative phosphorylation are coordinately upregulated with fasting hyperglycaemia in livers of patients with type 2 diabetes. Diabetologia. 2007;50(2):268–277. doi: 10.1007/s00125-006-0489-8. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

