RNA-Seq reveals a xenobiotic stress response in the soybean aphid, Aphis glycines, when fed aphid-resistant soybean
- PMID: 25399334
- PMCID: PMC4289043
- DOI: 10.1186/1471-2164-15-972
RNA-Seq reveals a xenobiotic stress response in the soybean aphid, Aphis glycines, when fed aphid-resistant soybean
Abstract
Background: While much recent research has expanded our understanding of the molecular interactions between aphids and their host plants, it is lacking for the soybean aphid, Aphis glycines. Since its North American invasion, A. glycines has become one of the most damaging insect pests on this important crop. Five soybean genes for host plant resistance to A. glycines have been identified, but populations of A. glycines have already adapted to overcome these resistance genes. Understanding the molecular interactions between resistant soybean and A. glycines can provide clues to its adaptation mechanisms. Here, we used RNA-Sequencing to compare and contrast A. glycines gene expression when fed resistant (Rag1) and susceptible soybean.
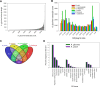
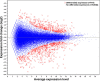
Results: Combining results from a previous A. glycines transcriptome, we generated 64,860 high quality transcripts, totaling 41,151,086 bases. Statistical analysis revealed 914 genes with significant differential expression. Most genes with higher expression in A. glycines on resistant plants (N = 352) were related to stress and detoxification such as cytochrome P450s, glutathione-S-transferases, carboxyesterases, and ABC transporters. A total of 562 genes showed lower transcript abundance in A. glycines on resistant plants. From our extensive transcriptome data, we also identified genes encoding for putative salivary effector proteins (N = 73). Among these, 6 effector genes have lower transcript abundance in A. glycines feeding on resistant soybean.
Conclusions: Overall, A. glycines exhibited a pattern typical of xenobiotic challenge, thereby validating antibiosis in Rag1, presumably mediated through toxic secondary metabolites. Additionally, this study identified many A. glycines genes and gene families at the forefront of its molecular interaction with soybean. Further investigation of these genes in other biotypes may reveal adaptation mechanisms to resistant plants.
Figures



References
-
- Tilmon KJ. Specialization, Speciation, and Radiation: the Evolutionary Biology of Herbivorous Insects. Berkeley, CA: University of California Press; 2008. p. 360.
-
- Smith CM. Plant Resistance to Arthropods: Molecular and Conventional Approaches. AA Dordrecht, The Netherlands: Springer; 2005. p. 423.
-
- Van Emden HF, Harrington R. Aphids as Crop Pests/edited by Helmut F. van Emden and Richard Harrington. Oxfordshire, UK: CABI Publishing; 2007.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

