Landscape of target:guide homology effects on Cas9-mediated cleavage
- PMID: 25399416
- PMCID: PMC4267615
- DOI: 10.1093/nar/gku1102
Landscape of target:guide homology effects on Cas9-mediated cleavage
Abstract
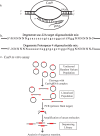
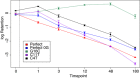
To study target sequence specificity, selectivity, and reaction kinetics of Streptococcus pyogenes Cas9 activity, we challenged libraries of random variant targets with purified Cas9::guide RNA complexes in vitro. Cleavage kinetics were nonlinear, with a burst of initial activity followed by slower sustained cleavage. Consistent with other recent analyses of Cas9 sequence specificity, we observe considerable (albeit incomplete) impairment of cleavage for targets mutated in the PAM sequence or in 'seed' sequences matching the proximal 8 bp of the guide. A second target region requiring close homology was located at the other end of the guide::target duplex (positions 13-18 relative to the PAM). Sequences flanking the guide+PAM region had measurable (albeit modest) effects on cleavage. In addition, the first-base Guanine constraint commonly imposed by gRNA expression systems has little effect on overall cleavage efficiency. Taken together, these studies provide an in vitro understanding of the complexities of Cas9-gRNA interaction and cleavage beyond the general paradigm of site determination based on the 'seed' sequence and PAM.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Westra E.R., Swarts D.C., Staals R.H.J., Jore M.M., Brouns S.J.J., van der Oost J. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 2012;46:311–339. - PubMed
-
- Bhaya D., Davison M., Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

