The OMA orthology database in 2015: function predictions, better plant support, synteny view and other improvements
- PMID: 25399418
- PMCID: PMC4383958
- DOI: 10.1093/nar/gku1158
The OMA orthology database in 2015: function predictions, better plant support, synteny view and other improvements
Abstract
The Orthologous Matrix (OMA) project is a method and associated database inferring evolutionary relationships amongst currently 1706 complete proteomes (i.e. the protein sequence associated for every protein-coding gene in all genomes). In this update article, we present six major new developments in OMA: (i) a new web interface; (ii) Gene Ontology function predictions as part of the OMA pipeline; (iii) better support for plant genomes and in particular homeologs in the wheat genome; (iv) a new synteny viewer providing the genomic context of orthologs; (v) statically computed hierarchical orthologous groups subsets downloadable in OrthoXML format; and (vi) possibility to export parts of the all-against-all computations and to combine them with custom data for 'client-side' orthology prediction. OMA can be accessed through the OMA Browser and various programmatic interfaces at http://omabrowser.org.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

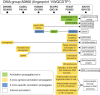
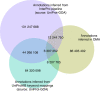
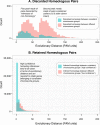

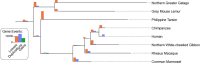
References
-
- Fitch W.M. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. - PubMed
-
- Sonnhammer E.L.L., Koonin E.V. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002;18:619–620. - PubMed
-
- Altenhoff A.M., Dessimoz C. Inferring orthology and paralogy. In: Anisimova M, editor. Evolutionary Genomics. Methods in Molecular Biology. Vol. 855. Clifton, NJ: Humana Press; 2012. pp. 259–279. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

