Introns and gene expression: cellular constraints, transcriptional regulation, and evolutionary consequences
- PMID: 25400101
- PMCID: PMC4654234
- DOI: 10.1002/bies.201400138
Introns and gene expression: cellular constraints, transcriptional regulation, and evolutionary consequences
Abstract
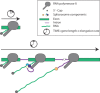
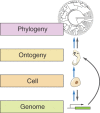
A gene's "expression profile" denotes the number of transcripts present relative to all other transcripts. The overall rate of transcript production is determined by transcription and RNA processing rates. While the speed of elongating RNA polymerase II has been characterized for many different genes and organisms, gene-architectural features - primarily the number and length of exons and introns - have recently emerged as important regulatory players. Several new studies indicate that rapidly cycling cells constrain gene-architecture toward short genes with a few introns, allowing efficient expression during short cell cycles. In contrast, longer genes with long introns exhibit delayed expression, which can serve as timing mechanisms for patterning processes. These findings indicate that cell cycle constraints drive the evolution of gene-architecture and shape the transcriptome of a given cell type. Furthermore, a tendency for short genes to be evolutionarily young hints at links between cellular constraints and the evolution of animal ontogeny.
Keywords: cell cycle constraints; gene length; macro-evolutionary patterns; splicing.
© 2015 The Authors. Bioessays published by WILEY Periodicals, Inc.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

