DNA polymerases engineered by directed evolution to incorporate non-standard nucleotides
- PMID: 25400626
- PMCID: PMC4215692
- DOI: 10.3389/fmicb.2014.00565
DNA polymerases engineered by directed evolution to incorporate non-standard nucleotides
Abstract
DNA polymerases have evolved for billions of years to accept natural nucleoside triphosphate substrates with high fidelity and to exclude closely related structures, such as the analogous ribonucleoside triphosphates. However, polymerases that can accept unnatural nucleoside triphosphates are desired for many applications in biotechnology. The focus of this review is on non-standard nucleotides that expand the genetic "alphabet." This review focuses on experiments that, by directed evolution, have created variants of DNA polymerases that are better able to accept unnatural nucleotides. In many cases, an analysis of past evolution of these polymerases (as inferred by examining multiple sequence alignments) can help explain some of the mutations delivered by directed evolution.
Keywords: AEGIS; CSR; DNA polymerases; directed evolution; non-standard nucleotides; protein engineering.
Figures
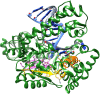
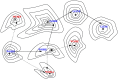
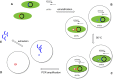
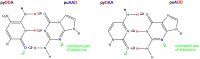
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

