Diverse allosteric and catalytic functions of tetrameric d-lactate dehydrogenases from three Gram-negative bacteria
- PMID: 25401076
- PMCID: PMC4230899
- DOI: 10.1186/s13568-014-0076-1
Diverse allosteric and catalytic functions of tetrameric d-lactate dehydrogenases from three Gram-negative bacteria
Abstract
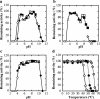
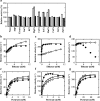
NAD-dependent d-lactate dehydrogenases (d-LDHs) reduce pyruvate into d-lactate with oxidation of NADH into NAD(+). Although non-allosteric d-LDHs from Lactobacilli have been extensively studied, the catalytic properties of allosteric d-LDHs from Gram-negative bacteria except for Escherichia coli remain unknown. We characterized the catalytic properties of d-LDHs from three Gram-negative bacteria, Fusobacterium nucleatum (FNLDH), Pseudomonas aeruginosa (PALDH), and E. coli (ECLDH) to gain an insight into allosteric mechanism of d-LDHs. While PALDH and ECLDH exhibited narrow substrate specificities toward pyruvate like usual d-LDHs, FNLDH exhibited a broad substrate specificity toward hydrophobic 2-ketoacids such as 2-ketobutyrate and 2-ketovalerate, the former of which gave a 2-fold higher k cat/S0.5 value than pyruvate. Whereas the three enzymes consistently showed hyperbolic shaped pyruvate saturation curves below pH 6.5, FNLDH and ECLDH, and PALDH showed marked positive and negative cooperativity, respectively, in the pyruvate saturation curves above pH 7.5. Oxamate inhibited the catalytic reactions of FNLDH competitively with pyruvate, and the PALDH reaction in a mixed manner at pH 7.0, but markedly enhanced the reactions of the two enzymes at low concentration through canceling of the apparent homotropic cooperativity at pH 8.0, although it constantly inhibited the ECLDH reaction. Fructose 1,6-bisphosphate and certain divalent metal ions such as Mg(2+) also markedly enhanced the reactions of FNLDH and PALDH, but none of them enhanced the reaction of ECLDH. Thus, our study demonstrates that bacterial d-LDHs have highly divergent allosteric and catalytic properties.
Keywords: Allosteric regulation; Escherichia coli; Fusobacterium nucleatum; Gram-negative bacteria; NAD-dependent d-lactate dehydrogenase; Pseudomonas aeruginosa.
Figures


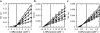

Similar articles
-
Structural Basis of Sequential Allosteric Transitions in Tetrameric d-Lactate Dehydrogenases from Three Gram-Negative Bacteria.Biochemistry. 2018 Sep 18;57(37):5388-5406. doi: 10.1021/acs.biochem.8b00557. Epub 2018 Sep 7. Biochemistry. 2018. PMID: 30149697
-
The Simple and Unique Allosteric Machinery of Thermus caldophilus Lactate Dehydrogenase : Structure-Function Relationship in Bacterial Allosteric LDHs.Adv Exp Med Biol. 2017;925:117-145. doi: 10.1007/5584_2016_171. Adv Exp Med Biol. 2017. PMID: 27815924 Review.
-
The archaeal LDH-like malate dehydrogenase from Ignicoccus islandicus displays dual substrate recognition, hidden allostery and a non-canonical tetrameric oligomeric organization.J Struct Biol. 2019 Oct 1;208(1):7-17. doi: 10.1016/j.jsb.2019.07.006. Epub 2019 Jul 10. J Struct Biol. 2019. PMID: 31301348
-
Deciphering Evolutionary Trajectories of Lactate Dehydrogenases Provides New Insights into Allostery.Mol Biol Evol. 2023 Oct 4;40(10):msad223. doi: 10.1093/molbev/msad223. Mol Biol Evol. 2023. PMID: 37797308 Free PMC article.
-
A molecular design that stabilizes active state in bacterial allosteric L-lactate dehydrogenases.J Biochem. 2011 Nov;150(5):579-91. doi: 10.1093/jb/mvr100. Epub 2011 Aug 9. J Biochem. 2011. PMID: 21828088
Cited by
-
D-Lactic Acid as a Metabolite: Toxicology, Diagnosis, and Detection.Biomed Res Int. 2020 Jun 17;2020:3419034. doi: 10.1155/2020/3419034. eCollection 2020. Biomed Res Int. 2020. PMID: 32685468 Free PMC article. Review.
-
Association between organic nitrogen substrates and the optical purity of D-lactic acid during the fermentation by Sporolactobacillus terrae SBT-1.Sci Rep. 2024 May 8;14(1):10522. doi: 10.1038/s41598-024-61247-4. Sci Rep. 2024. PMID: 38719898 Free PMC article.
-
A synthetic pathway for the production of 2-hydroxyisovaleric acid in Escherichia coli.J Ind Microbiol Biotechnol. 2018 Jul;45(7):579-588. doi: 10.1007/s10295-018-2005-9. Epub 2018 Jan 12. J Ind Microbiol Biotechnol. 2018. PMID: 29330665
-
Relative catalytic efficiencies and transcript levels of three d- and two l-lactate dehydrogenases for optically pure d-lactate production in Sporolactobacillus inulinus.Microbiologyopen. 2019 May;8(5):e00704. doi: 10.1002/mbo3.704. Epub 2018 Aug 1. Microbiologyopen. 2019. PMID: 30066438 Free PMC article.
-
Substrate Specificity and Allosteric Regulation of a D-Lactate Dehydrogenase from a Unicellular Cyanobacterium are Altered by an Amino Acid Substitution.Sci Rep. 2017 Nov 8;7(1):15052. doi: 10.1038/s41598-017-15341-5. Sci Rep. 2017. PMID: 29118438 Free PMC article.
References
-
- Arai K, Ishimitsu T, Fushinobu S, Uchikoba H, Matsuzawa H, Taguchi H. Active and inactive state structures of unliganded Lactobacillus casei allosteric l-lactate dehydrogenase. Proteins. 2010;78:681–694. doi: 10.1002/prot.22597. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

