Reconceptualizing the chlamydial inclusion as a pathogen-specified parasitic organelle: an expanded role for Inc proteins
- PMID: 25401095
- PMCID: PMC4215707
- DOI: 10.3389/fcimb.2014.00157
Reconceptualizing the chlamydial inclusion as a pathogen-specified parasitic organelle: an expanded role for Inc proteins
Abstract
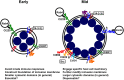
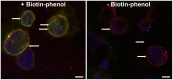
Chlamydia is an obligate intracellular pathogen that develops in the host cell in a vacuole termed the chlamydial inclusion. The prevailing concept of the chlamydial inclusion is of a parasitophorous vacuole. Here, the inclusion is the recipient of one-way host-pathogen interactions thus draining nutrients from the cell and negatively impacting it. While Chlamydia orchestrates some aspects of cell function, recent data indicate host cells remain healthy up until, and even after, chlamydial egress. Thus, while Chlamydia relies on the host cell for necessary metabolites, the overall function of the host cell, during chlamydial growth and development, is not grossly disturbed. This is consistent with the obligate intracellular organism's interest to maintain viability of its host. To this end, Chlamydia expresses inclusion membrane proteins, Incs, which serve as molecular markers for the inclusion membrane. Incs also contribute to the physical structure of the inclusion membrane and facilitate host-pathogen interactions across it. Given the function of Incs and the dynamic interactions that occur at the inclusion membrane, we propose that the inclusion behaves similarly to an organelle-albeit one that benefits the pathogen. We present the hypothesis that the chlamydial inclusion acts as a pathogen-specified parasitic organelle. This representation integrates the inclusion within existing subcellular trafficking pathways to divert a subset of host-derived metabolites thus maintaining host cell homeostasis. We review the known interactions of the chlamydial inclusion with the host cell and discuss the role of Inc proteins in the context of this model and how this perspective can impact the study of these proteins. Lessons learnt from the chlamydial pathogen-specified parasitic organelle can be applied to other intracellular pathogens. This will increase our understanding of how intracellular pathogens engage the host cell to establish their unique developmental niches.
Keywords: Chlamydia; inc protein; inclusion membrane; parasitophorous vacuole; pathogen specified organelle.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

