Specific genomic and transcriptomic aberrations in tumors induced by partial hepatectomy of a chronically inflamed murine liver
- PMID: 25401338
- PMCID: PMC4279375
- DOI: 10.18632/oncotarget.2515
Specific genomic and transcriptomic aberrations in tumors induced by partial hepatectomy of a chronically inflamed murine liver
Abstract
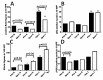
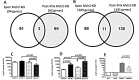
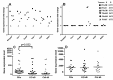
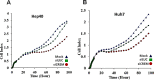
Resection of hepatocellular carcinoma (HCC) tumors by partial hepatectomy (PHx) is associated with promoting hepatocarcinogenesis. We have previously reported that PHx promotes hepatocarcinogenesis in the Mdr2-knockout (Mdr2-KO) mouse, a model for inflammation-mediated HCC. Now, to explore the molecular mechanisms underlying the tumor-promoting effect of PHx, we compared genomic and transcriptomic profiles of HCC tumors developing in the Mdr2-KO mice either spontaneously or following PHx. PHx accelerated HCC development in these mice by four months. PHx-induced tumors had major chromosomal aberrations: all were amplifications affecting multiple chromosomes. Most of these amplifications were located near the acrocentric centromeres of murine chromosomes. Four different chromosomal regions were amplified each in at least three tumors. The human orthologs of these common amplified regions are known to be amplified in HCC. All tumors of untreated mice had chromosomal aberrations, including both deletions and amplifications. Amplifications in spontaneous tumors affected fewer chromosomes and were not located preferentially at the chromosomal edges. Comparison of gene expression profiles revealed a significantly enriched expression of oncogenes, chromosomal instability markers and E2F1 targets in the post-PHx compared to spontaneous tumors. Both tumor groups shared the same frequent amplification at chromosome 18. Here, we revealed that one of the regulatory genes encoded by this amplified region, Crem, was over-expressed in the nuclei of murine and human HCC cells in vivo, and that it stimulated proliferation of human HCC cells in vitro. Our results demonstrate that PHx of a chronically inflamed liver directed tumor development to a discrete pathway characterized by amplification of specific chromosomal regions and expression of specific tumor-promoting genes. Crem is a new candidate HCC oncogene frequently amplified in this model and frequently over-expressed in human HCC.
Conflict of interest statement
Nothing to disclose.
Figures




References
-
- Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379(9822):1245–1255. - PubMed
-
- Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30(7):1073–1081. - PubMed
-
- Mauad TH, van Nieuwkerk CM, Dingemans KP, Smit JJ, Schinkel AH, Notenboom RG, van den Bergh Weerman MA, Verkruisen RP, Groen AK, Oude Elferink RPJ, van der Valk MA, Borst P, Offerhaus GJA. Mice with homozygous disruption of the mdr2 P-glycoprotein gene. A novel animal model for studies of nonsuppurative inflammatory cholangitis and hepatocarcinogenesis. Am J Pathol. 1994;145(5):1237–1245. - PMC - PubMed
-
- Katzenellenbogen M, Mizrahi L, Pappo O, Klopstock N, Olam D, Jacob-Hirsch J, Amariglio N, Rechavi G, Domany E, Galun E, Goldenberg D. Molecular mechanisms of liver carcinogenesis in the Mdr2-knockout mice. Mol Cancer Res. 2007;5(11):1159–1170. - PubMed
-
- Barash H, Gross ER, Edrei Y, Ella E, Israel A, Cohen I, Corchia N, Ben-Moshe T, Pappo O, Pikarsky E, Goldenberg D, Shiloh Y, Galun E, et al. Accelerated carcinogenesis following liver regeneration is associated with chronic inflammation-induced double-strand DNA breaks. Proc Natl Acad Sci U S A. 2010;107(5):2207–2212. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

