Cell fate decisions in malignant hematopoiesis: leukemia phenotype is determined by distinct functional domains of the MN1 oncogene
- PMID: 25401736
- PMCID: PMC4234417
- DOI: 10.1371/journal.pone.0112671
Cell fate decisions in malignant hematopoiesis: leukemia phenotype is determined by distinct functional domains of the MN1 oncogene
Abstract
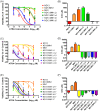
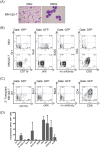
Extensive molecular profiling of leukemias and preleukemic diseases has revealed that distinct clinical entities, like acute myeloid (AML) and T-lymphoblastic leukemia (T-ALL), share similar pathogenetic mutations. It is not well understood how the cell of origin, accompanying mutations, extracellular signals or structural differences in a mutated gene determine the phenotypic identity of leukemias. We dissected the functional aspects of different protein regions of the MN1 oncogene and their effect on the leukemic phenotype, building on the ability of MN1 to induce leukemia without accompanying mutations. We found that the most C-terminal region of MN1 was required to block myeloid differentiation at an early stage, and deletion of an extended C-terminal region resulted in loss of myeloid identity and cell differentiation along the T-cell lineage in vivo. Megakaryocytic/erythroid lineage differentiation was blocked by the N-terminal region. In addition, the N-terminus was required for proliferation and leukemogenesis in vitro and in vivo through upregulation of HoxA9, HoxA10 and Meis2. Our results provide evidence that a single oncogene can modulate cellular identity of leukemic cells based on its active gene regions. It is therefore likely that different mutations in the same oncogene may impact cell fate decisions and phenotypic appearance of malignant diseases.
Conflict of interest statement
Figures




Similar articles
-
Extrinsic signals determine myeloid-erythroid lineage switch in MN1 leukemia.Exp Hematol. 2010 Mar;38(3):174-9. doi: 10.1016/j.exphem.2010.01.003. Epub 2010 Jan 21. Exp Hematol. 2010. PMID: 20096329
-
Mapping of MN1 sequences necessary for myeloid transformation.PLoS One. 2013 Apr 23;8(4):e61706. doi: 10.1371/journal.pone.0061706. Print 2013. PLoS One. 2013. PMID: 23626719 Free PMC article.
-
Meis2 as a critical player in MN1-induced leukemia.Blood Cancer J. 2017 Sep 29;7(9):e613. doi: 10.1038/bcj.2017.86. Blood Cancer J. 2017. PMID: 28960191 Free PMC article.
-
Differentiation primary response genes and proto-oncogenes as positive and negative regulators of terminal hematopoietic cell differentiation.Stem Cells. 1994 Jul;12(4):352-69. doi: 10.1002/stem.5530120402. Stem Cells. 1994. PMID: 7951003 Review.
-
The AML1 gene: a transcription factor involved in the pathogenesis of myeloid and lymphoid leukemias.Haematologica. 1997 May-Jun;82(3):364-70. Haematologica. 1997. PMID: 9234595 Review.
Cited by
-
Gain-of-Function MN1 Truncation Variants Cause a Recognizable Syndrome with Craniofacial and Brain Abnormalities.Am J Hum Genet. 2020 Jan 2;106(1):13-25. doi: 10.1016/j.ajhg.2019.11.011. Epub 2019 Dec 12. Am J Hum Genet. 2020. PMID: 31839203 Free PMC article.
-
MN1 Neurodevelopmental Disease-Atypical Phenotype Due to a Novel Frameshift Variant in the MN1 Gene.Front Mol Neurosci. 2021 Dec 16;14:789778. doi: 10.3389/fnmol.2021.789778. eCollection 2021. Front Mol Neurosci. 2021. PMID: 34975401 Free PMC article.
-
Chromosome 22q12.1 microdeletions: confirmation of the MN1 gene as a candidate gene for cleft palate.Eur J Hum Genet. 2016 Jan;24(1):51-8. doi: 10.1038/ejhg.2015.65. Epub 2015 May 6. Eur J Hum Genet. 2016. PMID: 25944382 Free PMC article.
-
Acquisition of an oncogenic fusion protein is sufficient to globally alter the landscape of miRNA expression to inhibit myogenic differentiation.Oncotarget. 2017 Jul 29;8(50):87054-87072. doi: 10.18632/oncotarget.19693. eCollection 2017 Oct 20. Oncotarget. 2017. PMID: 29152063 Free PMC article.
-
Role of Meningioma 1 for maintaining the transformed state in MLL-rearranged acute myeloid leukemia: potential for therapeutic intervention?Haematologica. 2020 May;105(5):1174-1176. doi: 10.3324/haematol.2019.246348. Haematologica. 2020. PMID: 32358078 Free PMC article. No abstract available.
References
-
- Gilliland DG (2001) Hematologic malignancies. Curr Opin Hematol 8: 189–191. - PubMed
-
- Dick JE (2008) Stem cell concepts renew cancer research. Blood 112: 4793–4807. - PubMed
-
- Kirstetter P, Schuster MB, Bereshchenko O, Moore S, Dvinge H, et al. (2008) Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell 13: 299–310. - PubMed
-
- Deshpande AJ, Cusan M, Rawat VP, Reuter H, Krause A, et al. (2006) Acute myeloid leukemia is propagated by a leukemic stem cell with lymphoid characteristics in a mouse model of CALM/AF10-positive leukemia. Cancer Cell 10: 363–374. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

