Tunable protein degradation in bacteria
- PMID: 25402616
- PMCID: PMC4262603
- DOI: 10.1038/nbt.3053
Tunable protein degradation in bacteria
Abstract
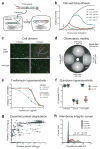
Tunable control of protein degradation in bacteria would provide a powerful research tool. Here we use components of the Mesoplasma florum transfer-messenger RNA system to create a synthetic degradation system that provides both independent control of steady-state protein level and inducible degradation of targeted proteins in Escherichia coli. We demonstrate application of this system in synthetic circuit development and control of core bacterial processes and antibacterial targets, and we transfer the system to Lactococcus lactis to establish its broad functionality in bacteria. We create a 238-member library of tagged essential proteins in E. coli that can serve as both a research tool to study essential gene function and an applied system for antibiotic discovery. Our synthetic protein degradation system is modular, does not require disruption of host systems and can be transferred to diverse bacteria with minimal modification.
Conflict of interest statement
The authors have submitted a patent application related to the work described here.
Figures
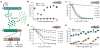


References
-
- Isaacs FJ, et al. Engineered riboregulators enable post-transcriptional control of gene expression. Nat Biotechnol. 2004;22:841–847. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

