A cell-based strategy to assess intrinsic inhibition efficiencies of HIV-1 reverse transcriptase inhibitors
- PMID: 25403670
- PMCID: PMC4335843
- DOI: 10.1128/AAC.04163-14
A cell-based strategy to assess intrinsic inhibition efficiencies of HIV-1 reverse transcriptase inhibitors
Abstract
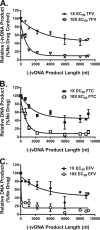
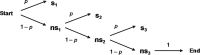
During HIV-1 reverse transcription, there are increasing opportunities for nucleos(t)ide (NRTI) or nonnucleoside (NNRTI) reverse transcriptase (RT) inhibitors to stop elongation of the nascent viral DNA (vDNA). In addition, RT inhibitors appear to influence the kinetics of vDNA synthesis differently. While cell-free kinetic inhibition constants have provided detailed mechanistic insight, these assays are dependent on experimental conditions that may not mimic the cellular milieu. Here we describe a novel cell-based strategy to provide a measure of the intrinsic inhibition efficiencies of clinically relevant RT inhibitors on a per-stop-site basis. To better compare inhibition efficiencies among HIV-1 RT inhibitors that can stop reverse transcription at any number of different stop sites, their basic probability, p, of getting stopped at any potential stop site was determined. A relationship between qPCR-derived 50% effective inhibitory concentrations (EC50s) and this basic probability enabled determination of p by successive approximation. On a per-stop-site basis, tenofovir (TFV) exhibited 1.4-fold-greater inhibition efficiency than emtricitabine (FTC), and as a class, both NRTIs exhibited an 8- to 11-fold greater efficiency than efavirenz (EFV). However, as more potential stops sites were considered, the probability of reverse transcription failing to reach the end of the template approached equivalence between both classes of RT inhibitors. Overall, this novel strategy provides a quantitative measure of the intrinsic inhibition efficiencies of RT inhibitors in the natural cellular milieu and thus may further understanding of drug efficacy. This approach also has applicability for understanding the impact of viral polymerase-based inhibitors (alone or in combination) in other virus systems.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Dead-end complexes contribute to the synergistic inhibition of HIV-1 RT by the combination of rilpivirine, emtricitabine, and tenofovir.Antiviral Res. 2014 Jan;101:131-5. doi: 10.1016/j.antiviral.2013.11.010. Epub 2013 Nov 28. Antiviral Res. 2014. PMID: 24291780
-
Biochemical and mechanistic basis for the activity of nucleoside analogue inhibitors of HIV reverse transcriptase.Curr Top Med Chem. 2004;4(10):1035-44. doi: 10.2174/1568026043388358. Curr Top Med Chem. 2004. PMID: 15193137 Review.
-
Novel nonnucleoside inhibitors that select nucleoside inhibitor resistance mutations in human immunodeficiency virus type 1 reverse transcriptase.Antimicrob Agents Chemother. 2006 Aug;50(8):2772-81. doi: 10.1128/AAC.00127-06. Antimicrob Agents Chemother. 2006. PMID: 16870771 Free PMC article.
-
Reduced emergence of the M184V/I resistance mutation when antiretroviral-naïve subjects use emtricitabine versus lamivudine in regimens composed of two NRTIs plus the NNRTI efavirenz.HIV Clin Trials. 2011 Mar-Apr;12(2):61-70. doi: 10.1310/hct1202-61. HIV Clin Trials. 2011. PMID: 21498149 Clinical Trial.
-
Intracellular interactions between nucleos(t)ide inhibitors of HIV reverse transcriptase.AIDS Rev. 2005 Apr-Jun;7(2):113-25. AIDS Rev. 2005. PMID: 16092505 Review.
Cited by
-
Determining 3'-Termini and Sequences of Nascent Single-Stranded Viral DNA Molecules during HIV-1 Reverse Transcription in Infected Cells.J Vis Exp. 2019 Jan 30;(143):10.3791/58715. doi: 10.3791/58715. J Vis Exp. 2019. PMID: 30774124 Free PMC article.
References
-
- Arts EJ, Stetor SR, Li X, Rausch JW, Howard KJ, Ehresmann B, North TW, Wohrl BM, Goody RS, Wainberg MA, Grice SF. 1996. Initiation of (−) strand DNA synthesis from tRNA(3Lys) on lentiviral RNAs: implications of specific HIV-1 RNA-tRNA(3Lys) interactions inhibiting primer utilization by retroviral reverse transcriptases. Proc Natl Acad Sci U S A 93:10063–10068. doi:10.1073/pnas.93.19.10063. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

