Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients
- PMID: 25403800
- PMCID: PMC4276431
- DOI: 10.1111/ajt.13031
Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients
Abstract
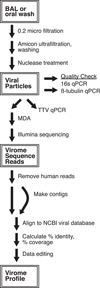
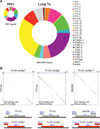
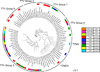
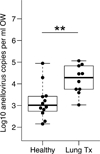
Few studies have examined the lung virome in health and disease. Outcomes of lung transplantation are known to be influenced by several recognized respiratory viruses, but global understanding of the virome of the transplanted lung is incomplete. To define the DNA virome within the respiratory tract following lung transplantation we carried out metagenomic analysis of allograft bronchoalveolar lavage (BAL), and compared with healthy and HIV+ subjects. Viral concentrates were purified from BAL and analyzed by shotgun DNA sequencing. All of the BAL samples contained reads mapping to anelloviruses, with high proportions in lung transplant samples. Anellovirus populations in transplant recipients were complex, with multiple concurrent variants. Quantitative polymerase chain reaction quantification revealed that anellovirus sequences were 56-fold more abundant in BAL from lung transplant recipients compared with healthy controls or HIV+ subjects (p < 0.0001). Anellovirus sequences were also more abundant in upper respiratory tract specimens from lung transplant recipients than controls (p = 0.006). Comparison to metagenomic data on bacterial populations showed that high anellovirus loads correlated with dysbiotic bacterial communities in allograft BAL (p = 0.008). Thus the respiratory tracts of lung transplant recipients contain high levels and complex populations of anelloviruses, warranting studies of anellovirus lung infection and transplant outcome.
Keywords: immunosuppression/immune modulation; lung transplantation/pulmonology; lung transplantation: living donor; microbiomics; molecular biology; translational research/science.
© Copyright 2014 The American Society of Transplantation and the American Society of Transplant Surgeons.
Figures



References
-
- Vu DL, Bridevaux PO, Aubert JD, Soccal PM, Kaiser L. Respiratory viruses in lung transplant recipients: A critical review and pooled analysis of clinical studies. American journal of transplantation: official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2011;11:1071–1078. - PMC - PubMed
-
- Garbino J, Gerbase MW, Wunderli W, Kolarova L, Nicod LP, Rochat T, Kaiser L. Respiratory viruses and severe lower respiratory tract complications in hospitalized patients. Chest. 2004;125:1033–1039. - PubMed
-
- Clark NM, Lynch JP, 3rd, Sayah D, Belperio JA, Fishbein MC, Weigt SS. DNA viral infections complicating lung transplantation. Seminars in respiratory and critical care medicine. 2013;34:380–404. - PubMed
-
- Speich R, van der Bij W. Epidemiology and management of infections after lung transplantation. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2001;33(Suppl 1):S58–S65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

