Long non-coding RNA HOTAIR is associated with human cervical cancer progression
- PMID: 25405331
- PMCID: PMC4277242
- DOI: 10.3892/ijo.2014.2758
Long non-coding RNA HOTAIR is associated with human cervical cancer progression
Abstract
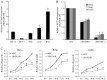
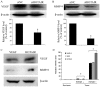
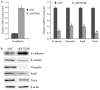
The functions of many long non-coding RNAs (lncRNAs) in human cancers remain to be clarified. The lncRNA Hox transcript antisense intergenic RNA (HOTAIR) has been reported to reprogram chromatin organization and promote breast and colorectal cancer metastasis, the involvement of lncRNAs in cervical cancer is just beginning to be studied. In the present study, we examined the expression and the functional role of HOTAIR in cervical cancer. HOTAIR expression was determined in cervical cancer tissues (n=111) and corresponding normal tissues (n=40) by using real-time polymerase chain reaction, and its correlation with clinical parameters and prognosis were analyzed. To determine the effect of HOTAIR knockdown and overexpression in cervical cancer cell lines, we used the CCK-8 assay, wound healing migration and matrigel invasion assay. The expression level of HOTAIR in cervical cancer tissues was higher than that in corresponding non-cancerous tissues. High HOTAIR expression correlated with lymph node metastasis, and reduced overall survival. A multivariate analysis showed that HOTAIR was a prognostic factor for predicting cervical cancer recurrence. Knockdown of HOTAIR reduced cell proliferation, migration, and invasion in cervical cancer cell lines. Moreover, HOTAIR regulated the expression of vascular endothelial growth factor, matrix metalloproteinase-9 and epithelial-to-mesenchymal transition (EMT)-related genes, which are important for cell motility and metastasis. Therefore, HOTAIR may promote tumor aggressiveness through the upregulation of VEGF and MMP-9 and EMT-related genes. These findings indicate that HOTAIR may represent a novel biomarker for predicting recurrence and prognosis and serve as a promising therapeutic target in cervical cancer.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

